Using GIS to Study Social Determinants of Health with R
Before we talk about code
Prerequisites
R
RStudio or other IDE
R packages (see Notes)
Prior knowledge of basic tidyverse functions.
If you are looking to learn how to use the R and the tidyverse for data science, we recommend R for Data Science. For more information about how R the programming language works, we recommend Advanced R.
Why are you here?
What questions led to you taking this training?
(Almost) All Data is Spatial
When conducting population research, almost everything has a spatial aspect to it.
We just might not have access to it
For those we do, it is important to incorporate spatial techniques to explore our data
Spatial Questions
How does a variable vary by region?
Is there a relationship between variables by region?
How many facilities are within a region?
How far is the nearest facility?
Does proximity to a facility have a relationship with another variable?
How has access changed over time?
Using R to Answer Spatial Questions
R is open source and reproducible
R is flexible
R can be used as part of a workflow or be the whole workflow

Working with Spatial Data
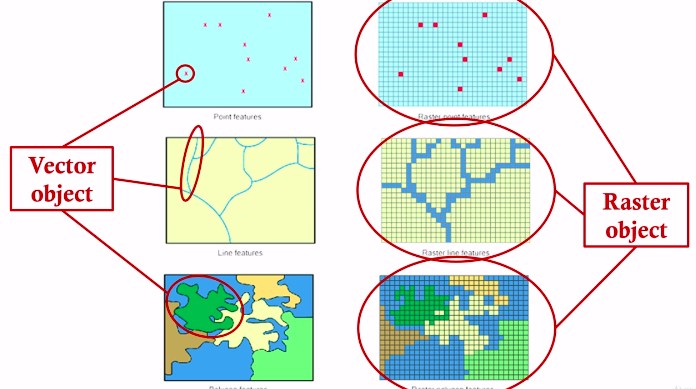
Vector vs Raster
- Vector data is made up of points, lines, and polygons
- sf package
- Raster data is made up of cells
- stars package
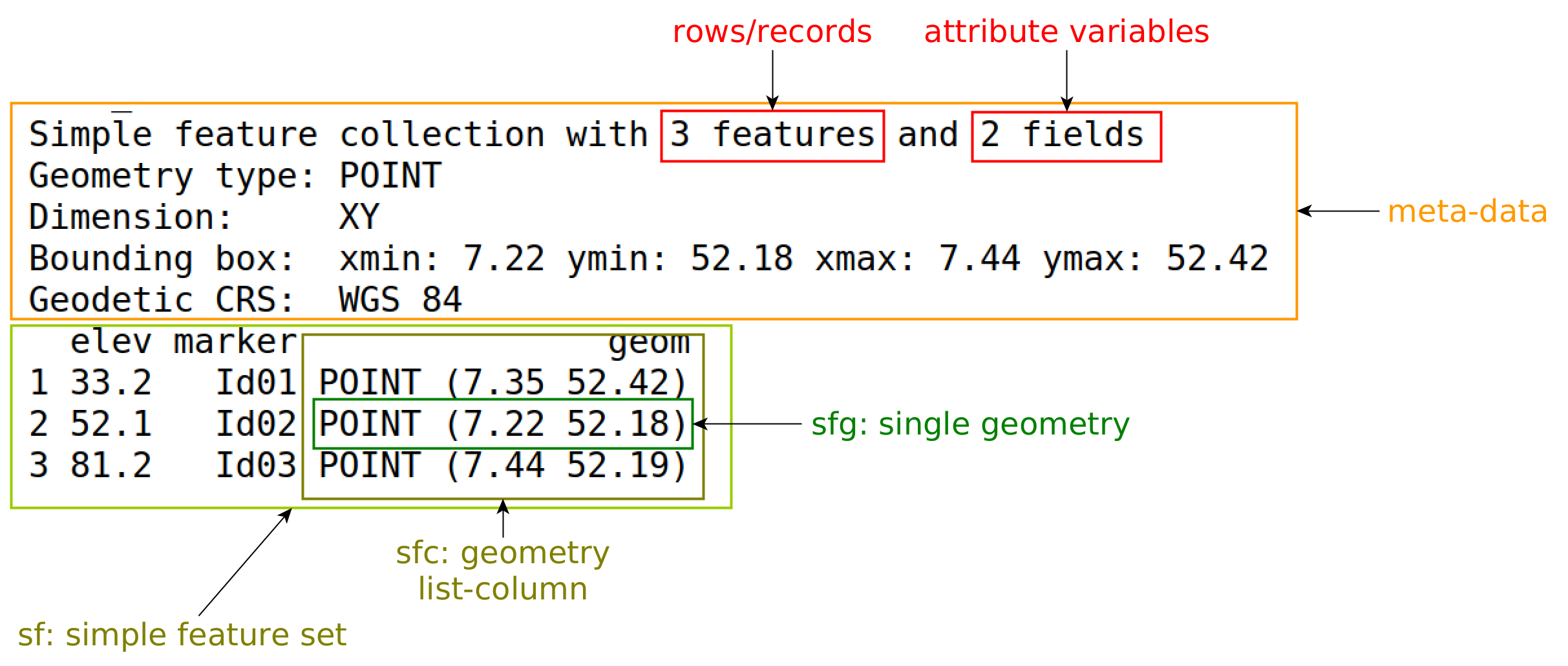
sf
- Primary package used for geocomputation within R
- Unified interface for several different geospatial libraries
- Represents spatial data as a
data.frameortibblewith a geometry list-column with the classsf
stars
- One of two modern packages used for rasters
- Allows for multidimensional data
- Well integrated with sf
- We will not cover most stars functionality

Spatial File Formats
| Shapefile | Geodatabase | Geojson | Geopackage | |
|---|---|---|---|---|
| Speed | Medium | Fast | Slow | Fast |
| Size limit | 2 GB | 256 TB | No limit | 140 TB |
| Files | At least 3 | Many | 1 | 1 |
| Multiple Features | No | Yes | No | Yes |
| Other Notes | Common | Proprietary | Plain Text | Open |
Reading Spatial Data in R
- Reading from sf is easy, simply specify the path
Simple feature collection with 157 features and 54 fields
Geometry type: POINT
Dimension: XY
Bounding box: xmin: -114.6334 ymin: 31.341 xmax: -109.04 ymax: 36.91705
Geodetic CRS: WGS 84
First 10 features:
OBJECTID OBJECTID_1 RUN_DATE source BUREAU FACID
1 1 2822 2024-05-06 ASPEN MED AZTH00002
2 2 2823 2024-05-06 ASPEN MED AZTH00003
3 3 2824 2024-05-06 ASPEN MED AZTH00004
4 4 3774 2024-05-06 ASPEN MED BH7347
5 5 4893 2024-05-06 ASPEN MED MED0198
6 6 4894 2024-05-06 ASPEN MED MED0201
7 7 4895 2024-05-06 ASPEN MED MED0203
8 8 4896 2024-05-06 ASPEN MED MED0204
9 9 4901 2024-05-06 ASPEN MED MED0209
10 10 4902 2024-05-06 ASPEN MED MED0211
FACILITY_N LICENSE_NU LICENSE_EF
1 BANNER- UNIVERSITY MEDICAL CENTER PHOENIX <NA> <NA>
2 BANNER UNIVERSITY MEDICAL CTR AT THE AZ HEALTH SC <NA> <NA>
3 MAYO CLINIC HOSPITAL <NA> <NA>
4 VIA LINDA BEHAVIORAL HOSPITAL SH11520 2023-03-01
5 CANYON VISTA MEDICAL CENTER H7130 2020-02-18
6 FLAGSTAFF MEDICAL CENTER H0169 2020-12-01
7 PAGE HOSPITAL H0086 2022-01-06
8 BANNER PAYSON MEDICAL CENTER H7250 2021-10-15
9 ABRAZO ARROWHEAD CAMPUS H0175 2021-12-01
10 BANNER BEHAVIORAL HEALTH HOSPITAL SH0147 2021-02-01
LICENSE_EX MEDICARE_I MEDICARE_1 MEDICARE_2 TELEPHONE FACILITY_T
1 <NA> 039802 <NA> <NA> (602)239-2716 NA
2 <NA> 039800 <NA> <NA> (520)694-0111 NA
3 <NA> 039801 <NA> <NA> (480)342-1900 NA
4 2025-02-28 034040 <NA> <NA> (480)476-7000 12
5 2024-12-27 030043 <NA> <NA> (520)263-2220 11
6 2024-11-30 030023 <NA> <NA> (928)779-3366 11
7 2024-12-28 031304 <NA> <NA> (928)645-2424 14
8 2024-12-24 031318 <NA> <NA> (928)471-3222 14
9 2024-12-27 030094 <NA> <NA> (623)561-1000 11
10 2024-12-24 034004 <NA> <NA> (480)448-7500 12
TYPE SUBTYPE CATEGORY ICON_CATEG
1 HOSPITAL TRANSPLANT HOSPITAL HOSPITAL
2 HOSPITAL TRANSPLANT HOSPITAL HOSPITAL
3 HOSPITAL TRANSPLANT HOSPITAL HOSPITAL
4 HOSPITAL PSYCHIATRIC HOSPITAL HOSPITAL
5 HOSPITAL SHORT TERM HOSPITAL HOSPITAL
6 HOSPITAL SHORT TERM HOSPITAL HOSPITAL
7 HOSPITAL CRITICAL ACCESS HOSPITALS HOSPITAL HOSPITAL
8 HOSPITAL CRITICAL ACCESS HOSPITALS HOSPITAL HOSPITAL
9 HOSPITAL SHORT TERM HOSPITAL HOSPITAL
10 HOSPITAL PSYCHIATRIC HOSPITAL HOSPITAL
MEDICARE_T LICENSE_TY LICENSE_SU
1 HOSPITAL - TRANSPLANT HOSPITAL FEDERAL ONLY <NA>
2 HOSPITAL - TRANSPLANT HOSPITAL FEDERAL ONLY <NA>
3 HOSPITAL - TRANSPLANT HOSPITAL FEDERAL ONLY <NA>
4 HOSPITAL - PSYCHIATRIC HOSPITAL HOSPITAL - SPECIAL
5 HOSPITAL - SHORT TERM HOSPITAL HOSPITAL - GENERAL
6 HOSPITAL - SHORT TERM HOSPITAL HOSPITAL - GENERAL
7 HOSPITAL - CRITICAL ACCESS HOSPITALS HOSPITAL HOSPITAL - GENERAL
8 HOSPITAL - CRITICAL ACCESS HOSPITALS HOSPITAL HOSPITAL - GENERAL
9 HOSPITAL - SHORT TERM HOSPITAL HOSPITAL - GENERAL
10 HOSPITAL - PSYCHIATRIC HOSPITAL HOSPITAL - SPECIAL
CAPACITY ADDRESS CITY ZIP COUNTY OPERATION_
1 0 1441 N 12TH PHOENIX 85006 MARICOPA ACTIVE
2 0 1501 NORTH CAMPBELL AVENUE TUCSON 85724 PIMA ACTIVE
3 0 5777 EAST MAYO BOULEVARD PHOENIX 85054 MARICOPA ACTIVE
4 120 9160 EAST HORSESHOE RD SCOTTSDALE 85258 MARICOPA ACTIVE
5 100 5700 EAST HIGHWAY 90 SIERRA VISTA 85635 COCHISE ACTIVE
6 268 1200 NORTH BEAVER STREET FLAGSTAFF 86001 COCONINO ACTIVE
7 25 501 NORTH NAVAJO DRIVE PAGE 86040 COCONINO ACTIVE
8 25 807 SOUTH PONDEROSA DRIVE PAYSON 85541 GILA ACTIVE
9 229 18701 NORTH 67TH AVENUE GLENDALE 85308 MARICOPA ACTIVE
10 156 7575 EAST EARLL DRIVE SCOTTSDALE 85251 MARICOPA ACTIVE
X_Is_Publi HOSPITAL_G KeyID ADHSCODE N_AddressT N_LocatorT
1 Yes <NA> AZTH00002 A AS0 CENTRUS-TOMTOM
2 Yes <NA> AZTH00003 A AS0 CENTRUS-TOMTOM
3 Yes <NA> AZTH00004 A AS0 CENTRUS-TOMTOM
4 Yes <NA> BH7347 A AS0 CENTRUS-TOMTOM
5 Yes <NA> MED0198 A AS0 CENTRUS-TOMTOM
6 Yes <NA> MED0201 A AS0 CENTRUS-TOMTOM
7 Yes <NA> MED0203 A AS0 CENTRUS-TOMTOM
8 Yes <NA> MED0204 A AS0 CENTRUS-TOMTOM
9 Yes <NA> MED0209 A AS0 CENTRUS-TOMTOM
10 Yes <NA> MED0211 A AS0 CENTRUS-TOMTOM
N_ADDRESS N_ADDR2 N_CITY N_COUNTY
1 1441 N 12TH ST <NA> PHOENIX MARICOPA COUNTY
2 1501 N CAMPBELL AVE <NA> TUCSON PIMA COUNTY
3 5777 E MAYO BLVD <NA> PHOENIX MARICOPA COUNTY
4 9160 E HORSESHOE RD <NA> SCOTTSDALE MARICOPA COUNTY
5 5700 E HIGHWAY 90 <NA> SIERRA VISTA COCHISE COUNTY
6 1200 N BEAVER ST <NA> FLAGSTAFF COCONINO COUNTY
7 501 N NAVAJO DR <NA> PAGE COCONINO COUNTY
8 807 S PONDEROSA ST <NA> PAYSON GILA COUNTY
9 18701 N 67TH AVE <NA> GLENDALE MARICOPA COUNTY
10 7575 E EARLL DR <NA> SCOTTSDALE MARICOPA COUNTY
N_FULLADDR N_LAT N_LON N_STATE
1 1441 N 12TH ST , PHOENIX, AZ 85006-2837 33.46410 -112.0562 AZ
2 1501 N CAMPBELL AVE , TUCSON, AZ 85724-0001 32.24080 -110.9443 AZ
3 5777 E MAYO BLVD , PHOENIX, AZ 85054-4502 33.66316 -111.9557 AZ
4 9160 E HORSESHOE RD , SCOTTSDALE, AZ 85258-4666 33.56670 -111.8840 AZ
5 5700 E HIGHWAY 90 , SIERRA VISTA, AZ 85635-9110 31.55449 -110.2319 AZ
6 1200 N BEAVER ST , FLAGSTAFF, AZ 86001-3118 35.20949 -111.6447 AZ
7 501 N NAVAJO DR , PAGE, AZ 86040-0959 36.91705 -111.4634 AZ
8 807 S PONDEROSA ST , PAYSON, AZ 85541-5542 34.23130 -111.3212 AZ
9 18701 N 67TH AVE , GLENDALE, AZ 85308-7100 33.65504 -112.2027 AZ
10 7575 E EARLL DR , SCOTTSDALE, AZ 85251-6915 33.48385 -111.9183 AZ
N_ZIP N_ZIP4 N_BLOCK RESERVATIO RESERV_ID R_METHOD
1 85006 2837 4.013113e+13 <NA> <NA> <NA>
2 85724 1 4.019002e+13 <NA> <NA> <NA>
3 85054 4502 4.013615e+13 <NA> <NA> <NA>
4 85258 4666 4.013941e+13 Salt River Reservation 3340F Coordinates
5 85635 9110 4.003002e+13 <NA> <NA> <NA>
6 86001 3118 4.005000e+13 <NA> <NA> <NA>
7 86040 959 4.005002e+13 <NA> <NA> <NA>
8 85541 5542 4.007001e+13 <NA> <NA> <NA>
9 85308 7100 4.013616e+13 <NA> <NA> <NA>
10 85251 6915 4.013218e+13 <NA> <NA> <NA>
oPCA oPCA_ID P_Method P_Reliable
1 CENTRAL CITY VILLAGE 39 Coordinates <NA>
2 TUCSON CENTRAL 107 Coordinates <NA>
3 DESERT VIEW VILLAGE 30 Coordinates <NA>
4 SALT RIVER PIMA-MARICOPA INDIAN COMMUNITY 62 Coordinates <NA>
5 SIERRA VISTA 123 Coordinates <NA>
6 FLAGSTAFF 10 Coordinates <NA>
7 PAGE 7 Coordinates <NA>
8 PAYSON 23 Coordinates <NA>
9 GLENDALE NORTH 56 Coordinates <NA>
10 SCOTTSDALE SOUTH 45 Coordinates <NA>
SOURCE_VW CASELOAD geometry
1 Tableau.vw_licensing_facilities <NA> POINT (-112.0562 33.46411)
2 Tableau.vw_licensing_facilities <NA> POINT (-110.9443 32.24081)
3 Tableau.vw_licensing_facilities <NA> POINT (-111.9557 33.66317)
4 Tableau.vw_licensing_facilities <NA> POINT (-111.884 33.56671)
5 Tableau.vw_licensing_facilities <NA> POINT (-110.2319 31.5545)
6 Tableau.vw_licensing_facilities <NA> POINT (-111.6447 35.20949)
7 Tableau.vw_licensing_facilities <NA> POINT (-111.4634 36.91705)
8 Tableau.vw_licensing_facilities <NA> POINT (-111.3212 34.2313)
9 Tableau.vw_licensing_facilities <NA> POINT (-112.2027 33.65505)
10 Tableau.vw_licensing_facilities <NA> POINT (-111.9183 33.48386)Reading Spatial Data in R
- If you are reading from a file format with multiple layers, specify the layer
Simple feature collection with 139 features and 6 fields
Geometry type: MULTIPOLYGON
Dimension: XY
Bounding box: xmin: -114.8163 ymin: 31.33234 xmax: -109.0452 ymax: 37.00372
Geodetic CRS: WGS 84
First 10 features:
CSA_ID CSA_NAME AREA_SQMILE POP2020 Shape__Area Shape__Length
1 1 Colorado City 5071.6400 8017 141389179634 1890568.8
2 10 Williams 4607.9690 10528 128462767671 2396775.1
3 100 San Luis 117.6636 37668 3280270845 307260.4
4 101 Gold Canyon 354.4739 15095 9882163593 502534.3
5 102 Florence 560.2777 37684 15619641198 848217.8
6 103 San Tan Valley 208.9516 114281 5825234969 348896.7
7 104 Saddlebrooke/Oracle 2165.1450 24960 60360749130 1342973.8
8 105 Maricopa 187.5613 63845 5228908746 352286.1
9 106 Coolidge 120.7299 17121 3365754248 306430.5
10 107 Casa Grande 825.9896 65107 23027262644 736741.1
SHAPE
1 MULTIPOLYGON (((-113.982 37...
2 MULTIPOLYGON (((-112.5724 3...
3 MULTIPOLYGON (((-114.625 32...
4 MULTIPOLYGON (((-111.04 33....
5 MULTIPOLYGON (((-111.0667 3...
6 MULTIPOLYGON (((-111.5609 3...
7 MULTIPOLYGON (((-110.4518 3...
8 MULTIPOLYGON (((-112.0127 3...
9 MULTIPOLYGON (((-111.5545 3...
10 MULTIPOLYGON (((-112.0303 3...Reading Spatial Data in R
- If you want your output to be a tibble, specify
as_tibble = TRUE
Simple feature collection with 518 features and 9 fields
Geometry type: POINT
Dimension: XY
Bounding box: xmin: -114.7864 ymin: 31.33913 xmax: -109.0569 ymax: 37.17732
Geodetic CRS: WGS 84
# A tibble: 518 × 10
OBJECTID VENDOR_NAME ADDRESS CITY STATE ZIPCODE PHONE STREET_ADDRESS2
<int> <chr> <chr> <chr> <chr> <int> <dbl> <chr>
1 1 FRY'S FOOD AND D… 2700 W… TEMPE ARIZ… 85282 6.02e9 <NA>
2 2 FRY'S FOOD AND D… 3036 E… PHOE… ARIZ… 85016 6.02e9 <NA>
3 3 FRY'S FOOD AND D… 1835 E… TEMPE ARIZ… 85283 4.81e9 <NA>
4 4 FRY'S FOOD AND D… 1100 S… COTT… ARIZ… 86326 9.29e9 BUILDING A
5 5 FRY'S FOOD AND D… 9401 E… TUCS… ARIZ… 85710 5.21e9 <NA>
6 6 FRY'S FOOD AND D… 520 E.… PHOE… ARIZ… 85040 6.02e9 <NA>
7 7 FRY'S FOOD AND D… 201 N.… FLAG… ARIZ… 86001 4.81e9 <NA>
8 8 FRY'S FOOD AND D… 950 W.… PRES… ARIZ… 86301 9.29e9 <NA>
9 9 FRY'S FOOD AND D… 2480 N… TUCS… ARIZ… 85712 5.20e9 <NA>
10 10 FRY'S FOOD AND D… 7455 W… PEOR… ARIZ… 85381 6.23e9 <NA>
# ℹ 508 more rows
# ℹ 2 more variables: FULLADDRESS <chr>, geometry <POINT [°]>Reading Spatial Data in R
- You can even read from a url
Simple feature collection with 273 features and 32 fields
Geometry type: POINT
Dimension: XY
Bounding box: xmin: -114.7862 ymin: 31.36436 xmax: -109.9916 ymax: 36.91674
Geodetic CRS: WGS 84
First 10 features:
OBJECTID_1 OBJECTID RECID NAME
1 1 1 1 BANNER URGENT CARE
2 2 2 2 BANNER URGENT CARE
3 3 3 3 SUMMIT HEALTHCARE URGENT CARE
4 4 4 4 CARBON HEALTH
5 5 5 5 DIGNITY HEALTH URGENT CARE- GILBERT
6 6 6 6 DIGNITY HEALTH URGENT CARE- AHWATUKEE
7 7 7 7 PHOENIX CHILDREN'S SPECIALTY&URGENT CARE,E VLY OTC
8 8 8 8 HAVASU PRIMARY CARE AND PEDIATRICS
9 9 9 9 CARBON HEALTH
10 10 10 10 PHOENIX CHILDREN'S SPECIALTY & URGENT CARE NW CTR
ADDRESS CITY ZIP TELEPHONE
1 35945 NORTH GARY ROAD SAN TAN VALLEY 85143 4808275750
2 21980 NORTH 83RD AVENUE PEORIA 85383 6234656375
3 4951 SOUTH WHITE MOUNTAIN ROAD BUILDING A SHOW LOW 85901 9285323926
4 1880 EAST TANGERINE, SUITE 100 ORO VALLEY 85755 5209007007
5 1501 NORTH GILBERT ROAD GILBERT 85234 4807283000
6 4545 EAST CHANDLER BOULEVARD PHOENIX 85044 4807283000
7 5131 EAST SOUTHERN AVENUE MESA 85206 4808335437
8 1799 NORTH KIOWA BOULEVARD, SUITE 104 LAKE HAVASU CITY 86403 9285051030
9 1040 SOUTH HARRISON ROAD, SUITE 120 TUCSON 85715 5209095221
10 20325 NORTH 51ST AVENUE SUITE 116 GLENDALE 85308 6239725437
Latitude Longitude objID KeyID ADHSCODE N_AddressType N_LocatorType
1 33.20571 -111.5823 1 1 A ROOFTOP GOOGLE
2 33.68333 -112.2379 2 2 A AS0 CENTRUS-TOMTOM
3 34.20512 -110.0204 3 3 A AS0 CENTRUS-TOMTOM
4 32.42697 -110.9438 4 4 A AS0 CENTRUS-TOMTOM
5 33.37658 -111.7884 5 5 A AS0 CENTRUS-TOMTOM
6 33.30504 -111.9851 6 6 A AS0 CENTRUS-TOMTOM
7 33.39367 -111.7207 7 7 A AS0 CENTRUS-TOMTOM
8 34.50317 -114.3512 8 8 A AS0 CENTRUS-TOMTOM
9 32.20775 -110.7900 9 9 A AS0 CENTRUS-TOMTOM
10 33.67027 -112.1691 10 10 A AS0 CENTRUS-TOMTOM
N_ADDRESS N_ADDR2 N_CITY N_COUNTY
1 35945 North Gary Road <NA> San Tan Valley PINAL COUNTY
2 21980 N 83RD AVE <NA> PEORIA MARICOPA COUNTY
3 4951 S WHITE MOUNTAIN RD BLDG A <NA> SHOW LOW NAVAJO COUNTY
4 1880 E TANGERINE RD STE 100 <NA> ORO VALLEY PIMA COUNTY
5 1501 N GILBERT RD <NA> GILBERT MARICOPA COUNTY
6 4545 E CHANDLER BLVD <NA> PHOENIX MARICOPA COUNTY
7 5131 E SOUTHERN AVE <NA> MESA MARICOPA COUNTY
8 1799 KIOWA AVE STE 104 <NA> LAKE HAVASU CITY MOHAVE COUNTY
9 1040 S HARRISON RD STE 120 <NA> TUCSON PIMA COUNTY
10 20325 N 51ST AVE STE 116 <NA> GLENDALE MARICOPA COUNTY
N_FULLADDR N_LAT
1 35945 N GARY RD , SAN TAN VALLEY, AZ 85143-5748 33.20571
2 21980 N 83RD AVE , PEORIA, AZ 85383-1850 33.68333
3 4951 S WHITE MOUNTAIN RD BLDG A , SHOW LOW, AZ 85901-7827 34.20512
4 1880 E TANGERINE RD STE 100 , ORO VALLEY, AZ 85755-6238 32.42697
5 1501 N GILBERT RD , GILBERT, AZ 85234-2390 33.37658
6 4545 E CHANDLER BLVD , PHOENIX, AZ 85048-7643 33.30504
7 5131 E SOUTHERN AVE , MESA, AZ 85206-2799 33.39367
8 1799 KIOWA AVE STE 104 , LAKE HAVASU CITY, AZ 86403-2867 34.50317
9 1040 S HARRISON RD STE 120 , TUCSON, AZ 85748-6601 32.20775
10 20325 N 51ST AVE STE 116 , GLENDALE, AZ 85308-5665 33.67027
N_LON N_STATE N_ZIP N_ZIP4 N_BLOCK RESERVATIO RESERV_ID R_METHOD
1 -111.5823 AZ 85143 5748 4.021000e+13 <NA> <NA> <NA>
2 -112.2379 AZ 85383 1850 4.013614e+13 <NA> <NA> <NA>
3 -110.0204 AZ 85901 7827 4.017962e+13 <NA> <NA> <NA>
4 -110.9438 AZ 85755 6238 4.019005e+13 <NA> <NA> <NA>
5 -111.7884 AZ 85234 2390 4.013423e+13 <NA> <NA> <NA>
6 -111.9851 AZ 85048 7643 4.013117e+13 <NA> <NA> <NA>
7 -111.7207 AZ 85206 2799 4.013423e+13 <NA> <NA> <NA>
8 -114.3512 AZ 86403 2867 4.015953e+13 <NA> <NA> <NA>
9 -110.7900 AZ 85748 6601 4.019004e+13 <NA> <NA> <NA>
10 -112.1691 AZ 85308 5665 4.013614e+13 <NA> <NA> <NA>
PCA_NAME PCA_ID PCA_PCT geometry
1 SAN TAN VALLEY 92 100 POINT (-111.5823 33.20571)
2 PEORIA NORTH 48 100 POINT (-112.2379 33.68333)
3 SHOW LOW 15 100 POINT (-110.0204 34.20512)
4 ORO VALLEY 100 100 POINT (-110.9438 32.42697)
5 GILBERT NORTH 74 100 POINT (-111.7884 33.37659)
6 AHWATUKEE FOOTHILLS VILLAGE 42 100 POINT (-111.9851 33.30504)
7 MESA EAST 68 100 POINT (-111.7207 33.39367)
8 LAKE HAVASU CITY 6 100 POINT (-114.3512 34.50318)
9 TUCSON EAST 110 100 POINT (-110.79 32.20775)
10 GLENDALE NORTH 56 100 POINT (-112.1691 33.67026)tigris
- Census boundaries are readily accessible through the tigris package
states(),counties(),tracts(),block_groups(),blocks(), etc.
tigris
- Just be careful to specify what boundaries you want
TIGER
cb = FALSE

Cartographic
cb = TRUE

Writing Spatial Data in R
- Writing spatial data is just as easy.
Visualizing Spatial Data
Projections
The world is not flat
Any representation of the earth in 2 dimensions cannot preserve both shape and area
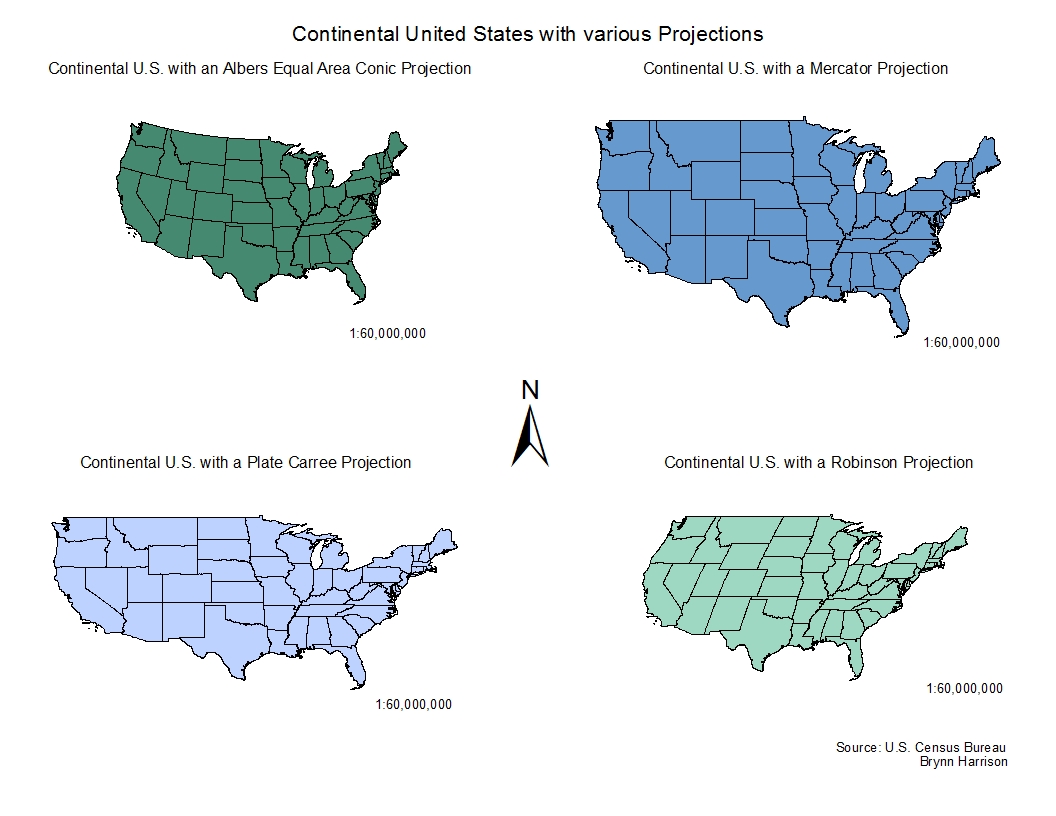
Projections
The projection you use will depend on the area of interest and your aim
The US Albers Equal Area Conic Projection is a standard for representing the lower 48
Each state has at least one state projection and the best one to may depend on who you ask
epsg.io is a good place to search for projections
![]()
Changing Projections
- To change a
sf’s projection usest_transform
plot
- The easiest way to look at any layer is using the plot function
- Note the use of
st_geometry, by default plot will try to construct maps of every attribute within thesfclass
plot
- The easiest way to look at any layer is using the plot function
- Note the use of
st_geometry, by default plot will try to construct maps of every attribute within thesfclass
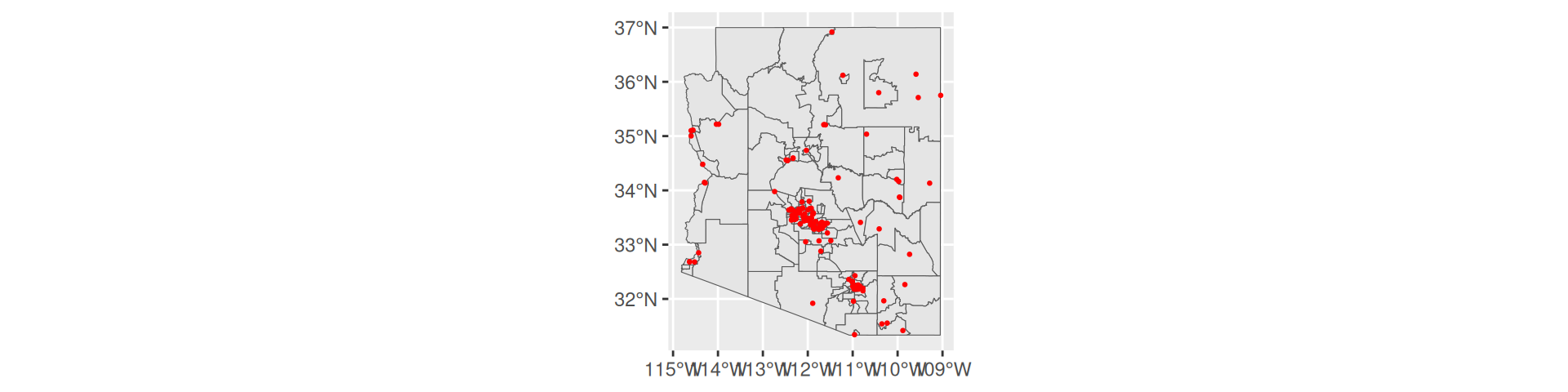
ggplot2
- Easy to use, same syntax as normal
- Much more customizable
leaflet
- Interactive
- More complex
leaflet
Stop and make a map
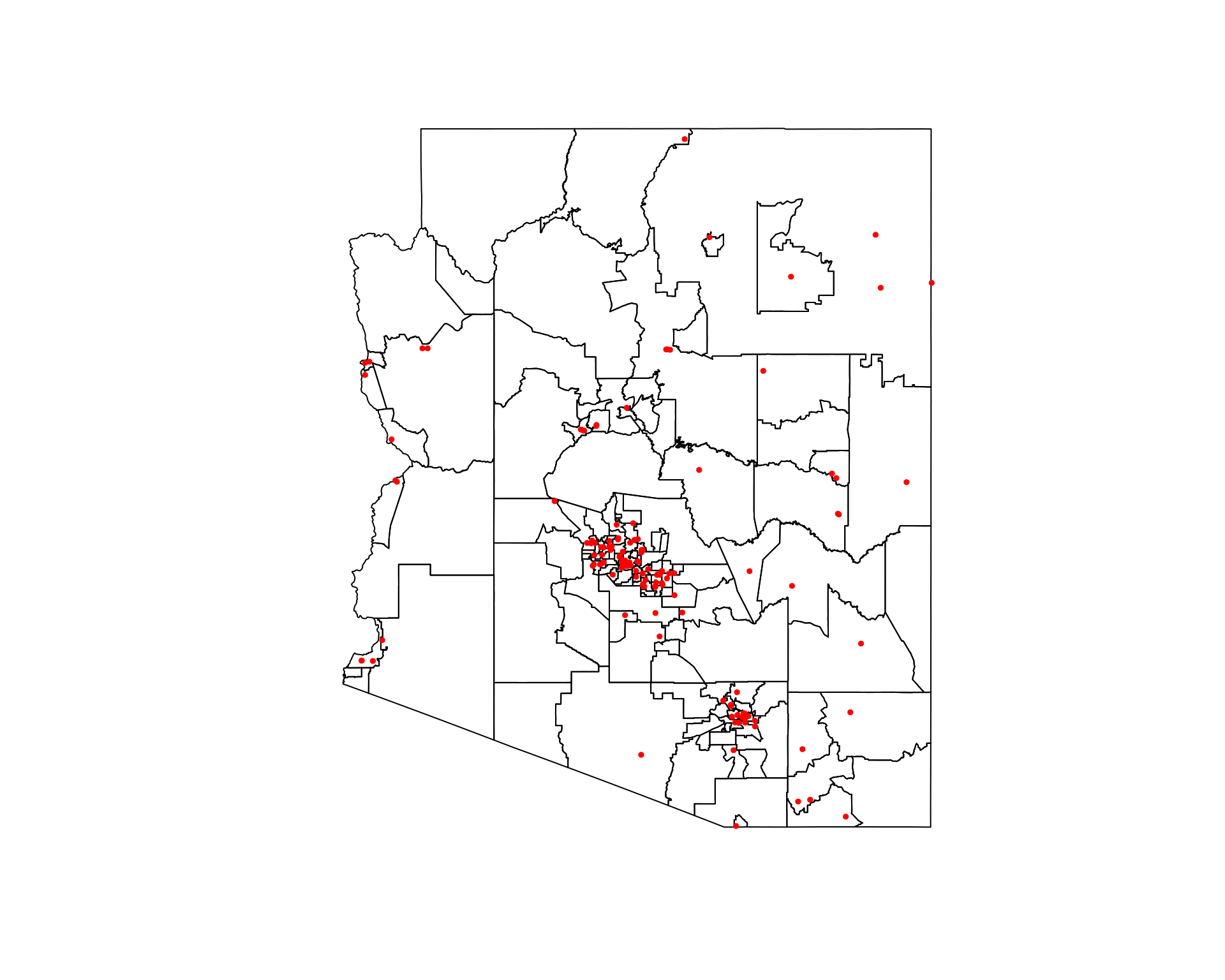
Use tigris to get county boundaries for the state of Arizona and overlay Arizona Hospitals (AZ_Hospitals.shp) within the Arizona Central projection (EPSG:26949) using ggplot2
Tips:
labs()- map title, legend title, legend labels
theme()andelement_text()- font, font size, color
theme_void()- remove long lat and background
Map Types
Choropleth
- Vary color by attribute
- Generalize data
- Useful for relative data
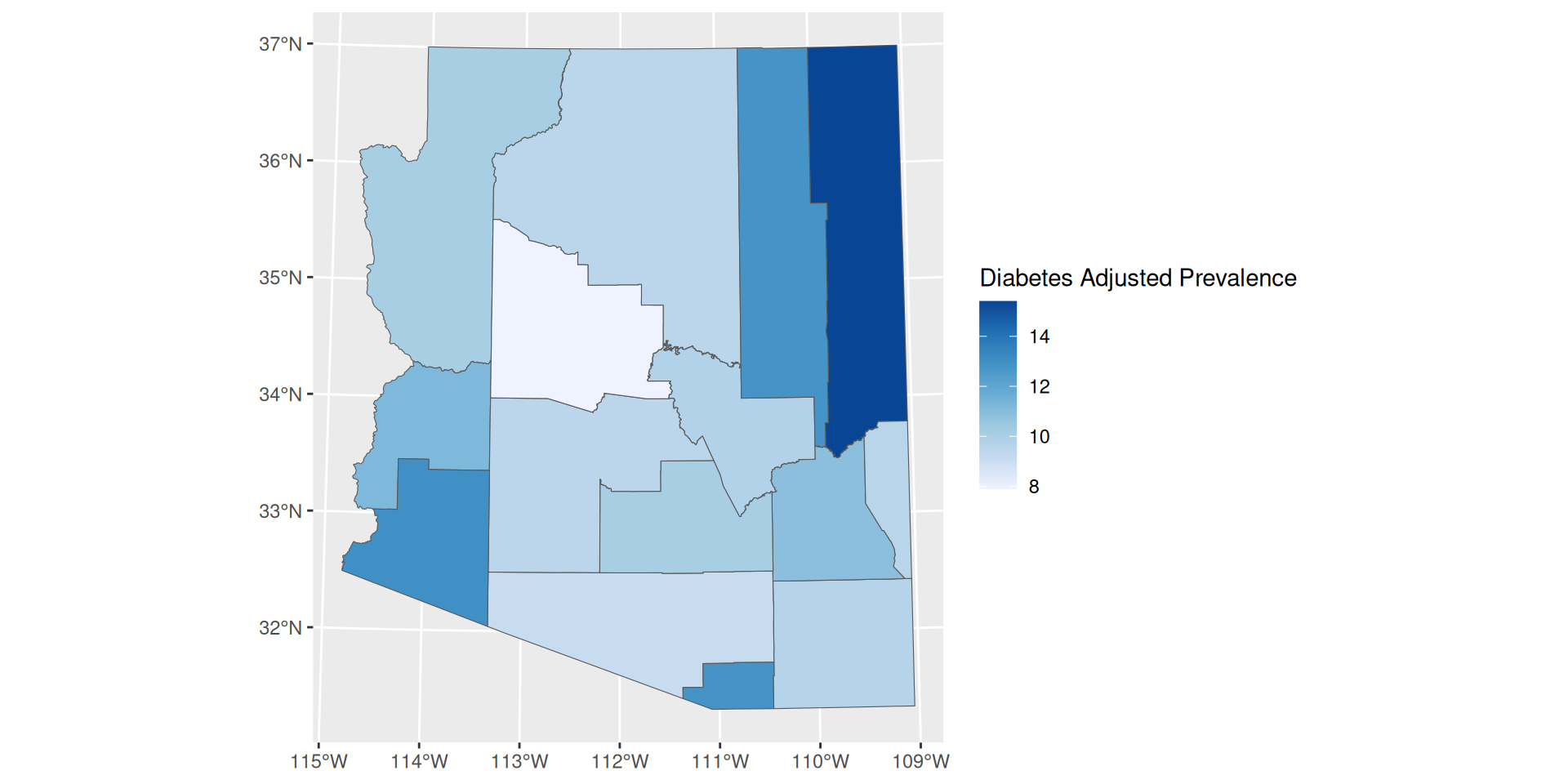
Continuous Scale
- Is default for ggplot
- Not robust to outliers
- Difficult to identify a color value and associate it with its value on the scale
scale_fill_distiller
Continuous Scale
Quantile Breaks
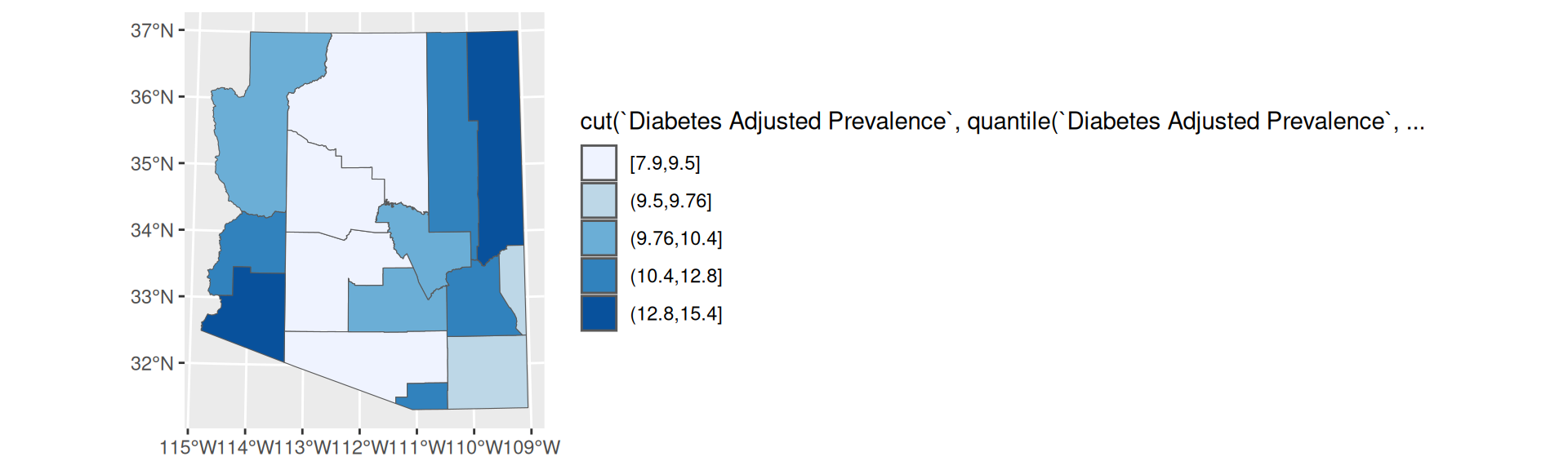
- Great for comparisons
- Can group regions that are very different
- Easy to identify a color value and associate it with its value on the scale
scale_fill_brewer
Quantile Breaks
Equal Interval Breaks
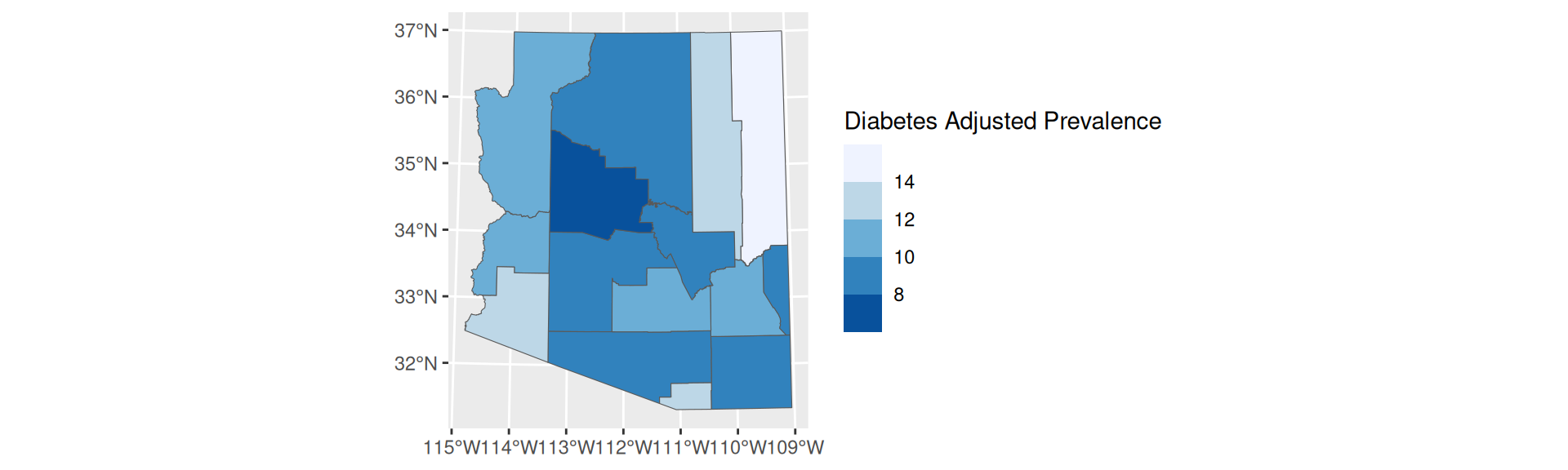
- Not robust to outliers
- Easy to identify a color value and associate it with its value on the scale
scale_fill_fermenter
Equal Interval Breaks
Manual Breaks
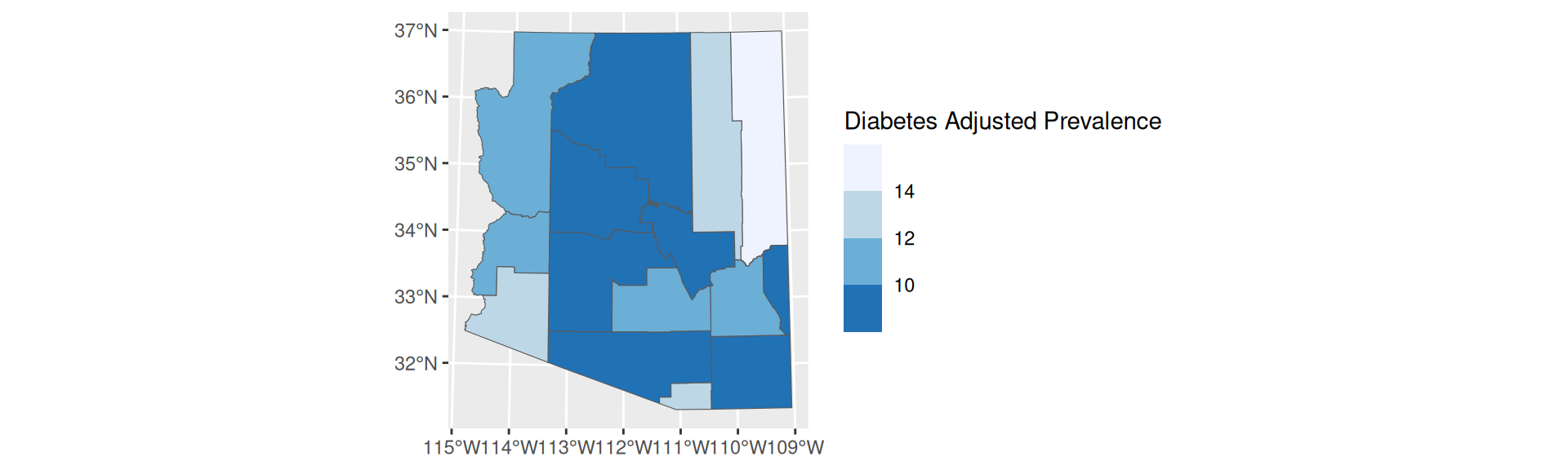
- Requires prior information about important breaks
- Easy to identify a color value and associate it with its value on the scale
scale_fill_fermenter
Manual Breaks
Cartograms
- Same as choropleth
- Transforms geographic areas so that their area is proportional to the population
- Types
- Continuous (
cartogram_cont) - distorts shape - Discontinuous (
cartogram_ncont) - maintains shape, distorts size - Dorling (
cartogram_dorling) - creates circles of weighted size
- Continuous (
Cartograms
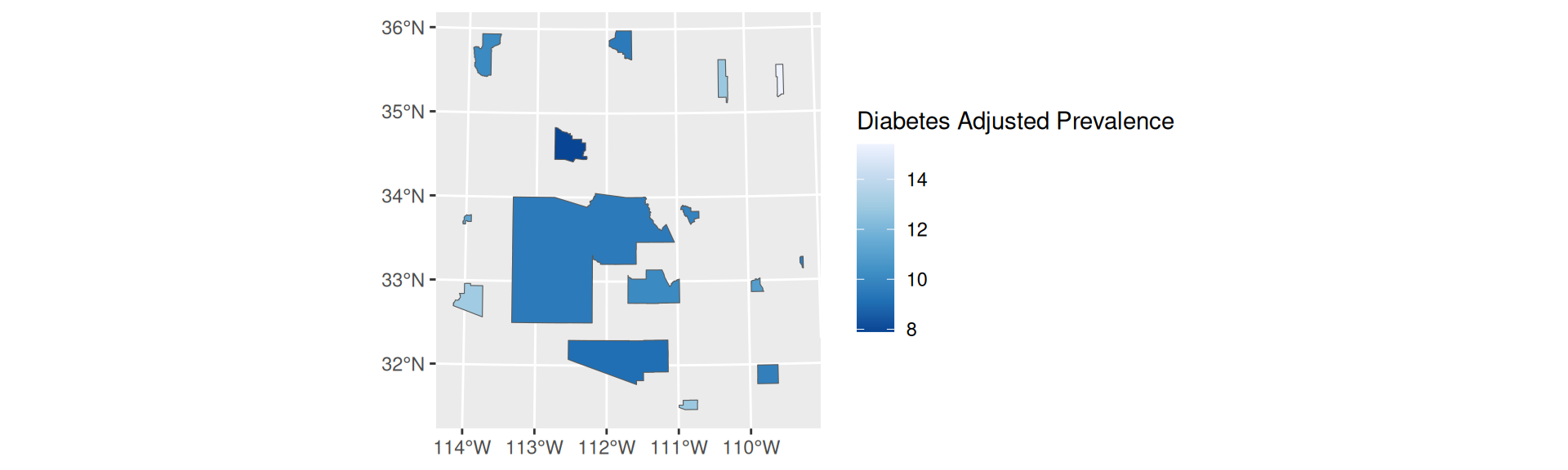
Proportional Symbol
- Choropleths are best used with relative data (aka rates)
- Symbol maps are best used with count data
- Like cartograms, symbol maps mitigate area bias
- Have a continuous scale
st_centroid,scale_size, andaes(pch = 20, size =FIELD)
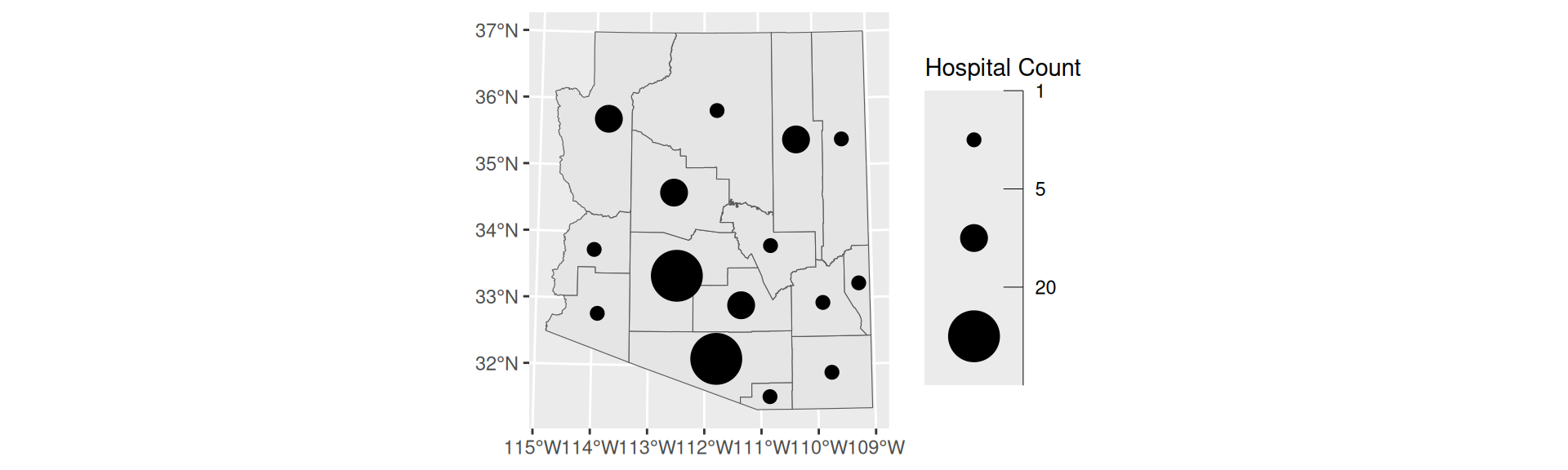
Proportional Symbol
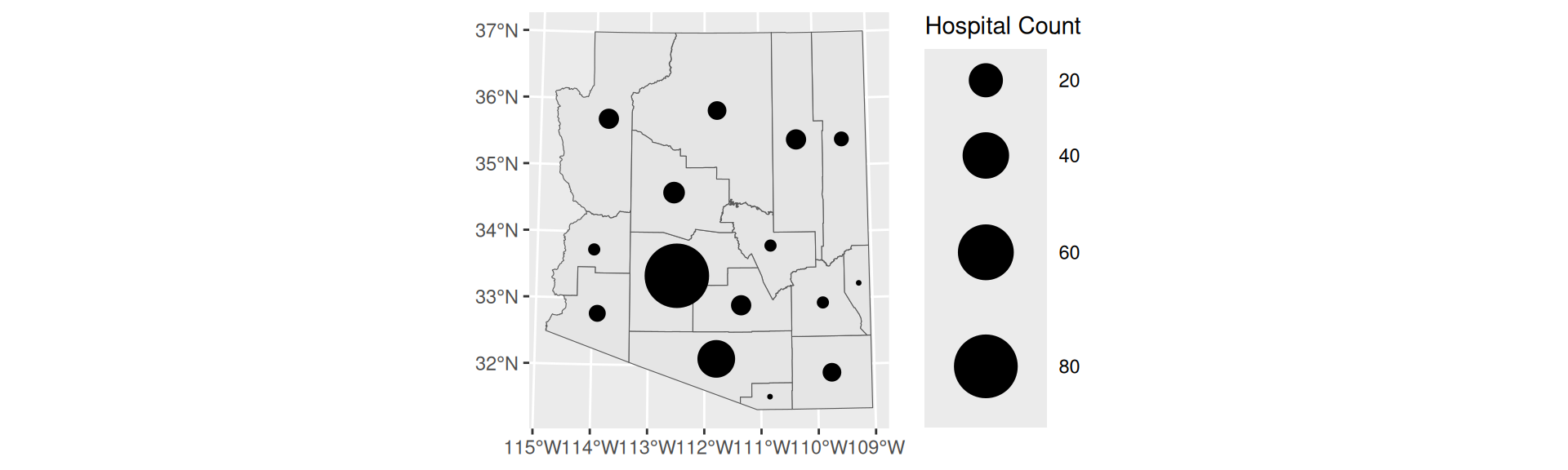
Graduated Symbol
- Choropleths are best used with relative data (aka rates)
- Symbol maps are best used with count data
- Like cartograms, symbol maps mitigate area bias
- Have binned values
st_centroid,scale_size_binned, andaes(pch = 20, size =FIELD)
Graduated Symbol
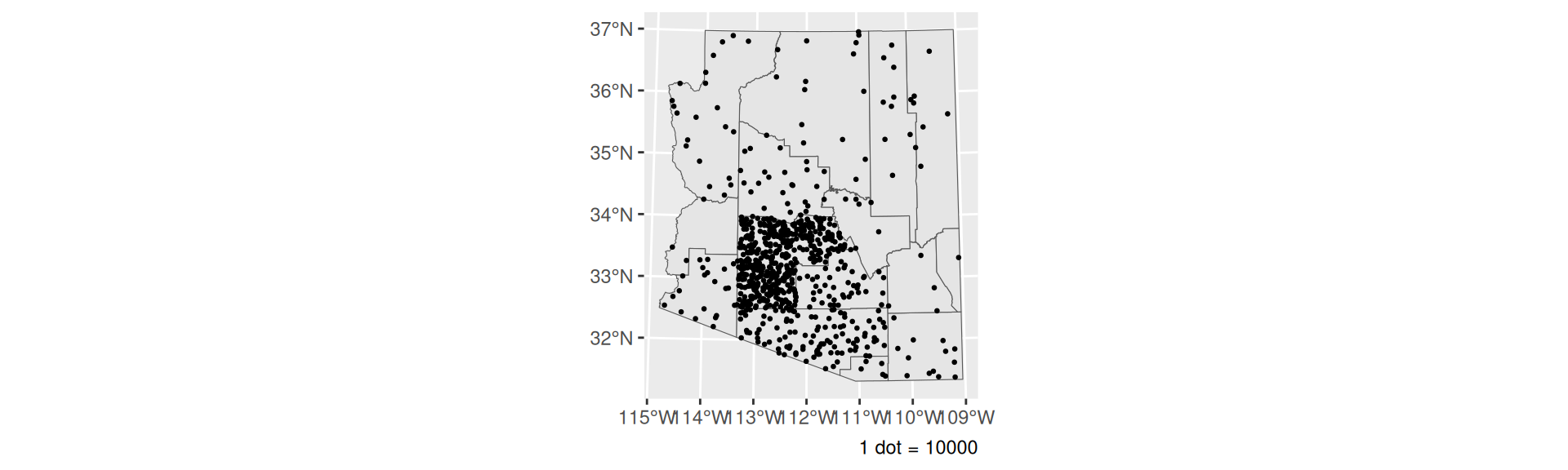
Dot Density
- Randomly distributed dots representing some number of counts
- Masks data
- From tidycensus,
as_dot_density
Dot Density
Stop and make some maps
- Make a quantile breaks choropleth map using the PLACES data (data/pres/sample.geojson) with hospital locations (data/AZ/hospitals) layered on top
- Alternatively, you can layer a symbol map on top
- Using
cowplot::plot_grid(plot1,plot2), plot two maps of different types, side by side
Geocomputation
More on Projections
- Much of the data you may come across will be unprojected
- In R, you can check if a
sfis unprojected by usingst_is_longlat()
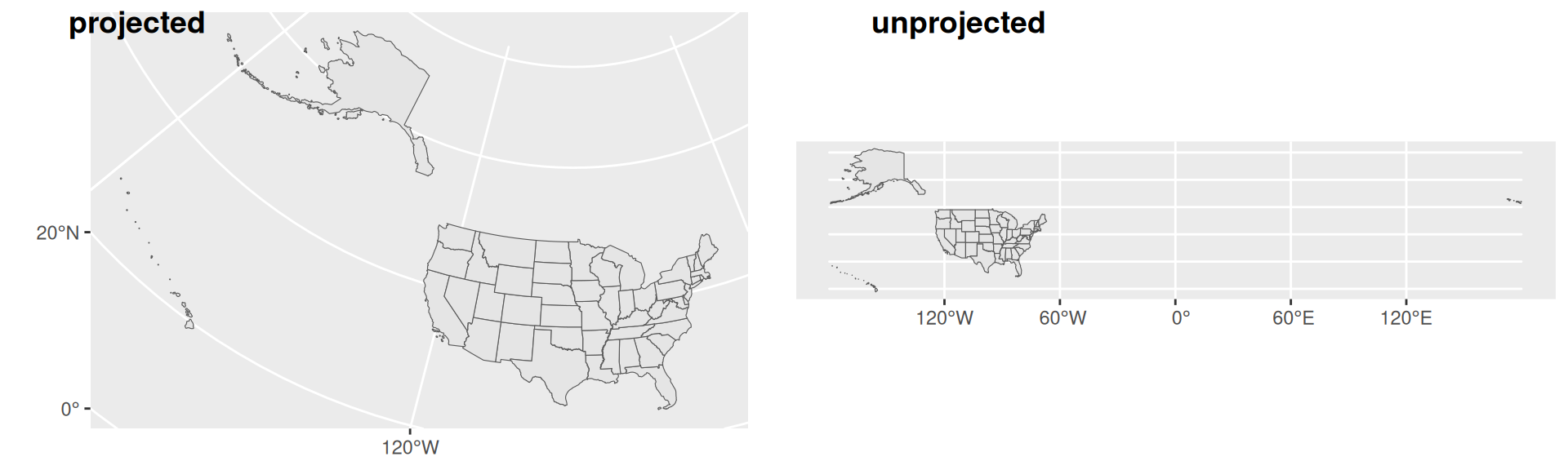
More on Projections
- R uses an geocomputing engine called S2 which represents the earth which approximates the earth as a sphere when using unprojected data
- S2 mitigates issues associated with assuming planar geometry
- S2 is not perfect, and often, using a good projection can reduce frustrations down the road
Typical Identifiers
For census geometries, we have FIPS codes (aka GEOIDs)
Area Structure Digits State STATE 2 County STATE+COUNTY 2+3=5 Tract STATE+COUNTY+TRACT 2+3+6=11 Block Group STATE+COUNTY+TRACT+BLOCK GROUP 2+3+6+1=12 Block STATE+COUNTY+TRACT+BLOCK 2+3+6+4=15
Tabular Joins
- Because
sfs are specialdata.framesortibbles, you can join data just as you would with a table. - The tables must have identical identifiers
- Just like table to table joins, you must be careful with data types.
left_join(),right_join(),inner_join(), andouter_join()all work
Tabular Joins
states <- counties(state = "AZ") %>%
select(GEOID)
PLACES <- read_csv("data/US/PLACES2023/PLACES2023_county.csv")
left_join(states, PLACES, join_by(GEOID == CountyFIPS))Simple feature collection with 15 features and 154 fields
Geometry type: MULTIPOLYGON
Dimension: XY
Bounding box: xmin: -114.8163 ymin: 31.33234 xmax: -109.0452 ymax: 37.00373
Geodetic CRS: NAD83
First 10 features:
GEOID StateAbbr StateDesc CountyName TotalPopulation ACCESS2_CrudePrev
1 04027 AZ Arizona Yuma 206990 23.6
2 04021 AZ Arizona Pinal 449557 14.1
3 04017 AZ Arizona Navajo 108147 14.6
4 04011 AZ Arizona Greenlee 9404 16.2
5 04013 AZ Arizona Maricopa 4496588 12.7
6 04019 AZ Arizona Pima 1052030 12.9
7 04003 AZ Arizona Cochise 126050 14.5
8 04005 AZ Arizona Coconino 145052 10.6
9 04007 AZ Arizona Gila 53589 12.9
10 04009 AZ Arizona Graham 39050 16.9
ACCESS2_Crude95CI ACCESS2_AdjPrev ACCESS2_Adj95CI ARTHRITIS_CrudePrev
1 (18.6, 29.5) 23.7 (18.8, 29.4) 22.9
2 (11.5, 17.0) 14.5 (11.8, 17.5) 28.1
3 (12.1, 17.7) 15.3 (12.7, 18.5) 29.8
4 (12.9, 19.4) 16.2 (13.0, 19.5) 21.2
5 (10.3, 15.3) 12.9 (10.4, 15.7) 22.9
6 (10.3, 16.1) 13.3 (10.6, 16.5) 26.3
7 (11.6, 17.5) 15.1 (12.1, 18.3) 29.3
8 ( 8.5, 13.3) 10.7 ( 8.7, 13.1) 18.9
9 (10.6, 15.5) 14.1 (11.6, 16.9) 31.6
10 (13.6, 20.3) 16.9 (13.7, 20.3) 24.2
ARTHRITIS_Crude95CI ARTHRITIS_AdjPrev ARTHRITIS_Adj95CI BINGE_CrudePrev
1 (20.0, 26.0) 20.0 (17.3, 22.9) 15.9
2 (25.2, 30.9) 23.5 (21.0, 26.2) 16.3
3 (26.7, 32.8) 24.9 (22.2, 27.6) 14.7
4 (17.8, 24.7) 20.1 (16.8, 23.5) 18.1
5 (20.9, 25.0) 21.1 (19.2, 23.0) 16.7
6 (23.9, 28.6) 22.8 (20.5, 24.9) 15.6
7 (26.2, 32.5) 23.5 (20.8, 26.3) 15.1
8 (16.7, 21.3) 20.0 (17.7, 22.6) 17.6
9 (28.2, 35.1) 22.3 (19.7, 25.0) 14.3
10 (20.7, 27.9) 23.6 (20.1, 27.3) 17.2
BINGE_Crude95CI BINGE_AdjPrev BINGE_Adj95CI BPHIGH_CrudePrev
1 (13.3, 18.7) 17.2 (14.4, 20.3) 33.3
2 (13.8, 18.8) 18.2 (15.5, 21.0) 33.0
3 (12.4, 17.1) 16.6 (14.0, 19.3) 37.3
4 (14.9, 21.7) 18.5 (15.2, 22.2) 27.0
5 (14.8, 18.6) 17.4 (15.5, 19.4) 29.6
6 (13.6, 17.8) 17.1 (15.0, 19.5) 31.7
7 (12.6, 17.8) 17.6 (14.8, 20.8) 36.5
8 (15.0, 20.5) 17.3 (14.8, 20.3) 25.1
9 (12.0, 16.7) 18.7 (15.7, 21.8) 37.0
10 (14.0, 20.6) 17.4 (14.2, 20.9) 30.1
BPHIGH_Crude95CI BPHIGH_AdjPrev BPHIGH_Adj95CI BPMED_CrudePrev
1 (29.9, 36.7) 30.2 (26.9, 33.6) 77.2
2 (29.9, 36.0) 28.5 (25.6, 31.4) 77.0
3 (33.9, 40.5) 32.3 (29.1, 35.3) 77.3
4 (23.3, 30.8) 25.8 (22.2, 29.5) 72.5
5 (27.3, 31.9) 27.7 (25.5, 29.9) 72.9
6 (29.0, 34.3) 28.1 (25.6, 30.5) 77.3
7 (33.2, 39.8) 30.4 (27.3, 33.6) 76.8
8 (22.5, 27.8) 26.6 (23.9, 29.6) 70.0
9 (33.6, 40.7) 27.3 (24.4, 30.3) 82.3
10 (26.3, 34.0) 29.5 (25.7, 33.4) 72.6
BPMED_Crude95CI BPMED_AdjPrev BPMED_Adj95CI CANCER_CrudePrev
1 (74.2, 79.8) 57.3 (52.7, 62.0) 6.9
2 (73.9, 79.7) 55.8 (51.4, 60.0) 7.8
3 (74.4, 80.0) 56.6 (52.3, 60.9) 7.7
4 (68.7, 76.0) 53.8 (48.6, 58.8) 6.1
5 (70.2, 75.3) 53.8 (50.5, 57.1) 6.9
6 (74.7, 79.7) 56.4 (52.7, 60.2) 7.6
7 (73.7, 79.7) 54.1 (49.8, 58.6) 8.1
8 (66.6, 73.1) 52.4 (48.2, 57.0) 5.9
9 (79.3, 84.7) 56.7 (52.0, 61.0) 10.1
10 (68.9, 76.1) 55.3 (50.0, 60.3) 6.2
CANCER_Crude95CI CANCER_AdjPrev CANCER_Adj95CI CASTHMA_CrudePrev
1 ( 6.2, 7.6) 5.5 ( 5.0, 6.1) 9.7
2 ( 7.0, 8.5) 6.1 ( 5.5, 6.7) 10.0
3 ( 6.9, 8.5) 6.1 ( 5.5, 6.7) 12.6
4 ( 5.5, 6.7) 5.7 ( 5.2, 6.3) 9.7
5 ( 6.3, 7.5) 6.2 ( 5.7, 6.8) 9.9
6 ( 6.9, 8.3) 6.1 ( 5.6, 6.7) 10.2
7 ( 7.4, 9.0) 6.0 ( 5.4, 6.6) 10.4
8 ( 5.4, 6.6) 6.3 ( 5.7, 6.9) 11.4
9 ( 9.1, 11.1) 6.4 ( 5.8, 7.1) 10.5
10 ( 5.6, 6.8) 5.9 ( 5.4, 6.6) 10.6
CASTHMA_Crude95CI CASTHMA_AdjPrev CASTHMA_Adj95CI CERVICAL_CrudePrev
1 ( 8.6, 11.0) 9.9 ( 8.7, 11.1) 76.4
2 ( 8.8, 11.2) 10.1 ( 8.9, 11.4) 79.4
3 (11.2, 14.0) 12.8 (11.4, 14.2) 74.7
4 ( 8.5, 11.0) 9.7 ( 8.5, 11.0) 78.5
5 ( 9.0, 11.0) 10.0 ( 9.0, 11.0) 80.6
6 ( 9.1, 11.4) 10.3 ( 9.2, 11.5) 78.1
7 ( 9.2, 11.7) 10.6 ( 9.4, 12.0) 79.0
8 (10.1, 12.8) 11.2 (10.0, 12.6) 75.3
9 ( 9.3, 11.8) 11.0 ( 9.7, 12.3) 77.1
10 ( 9.2, 12.1) 10.6 ( 9.2, 12.1) 75.4
CERVICAL_Crude95CI CERVICAL_AdjPrev CERVICAL_Adj95CI CHD_CrudePrev
1 (73.5, 79.0) 77.7 (75.1, 80.2) 7.6
2 (77.0, 81.3) 79.8 (77.7, 81.7) 7.0
3 (72.5, 76.9) 75.7 (73.5, 77.8) 9.0
4 (76.3, 80.8) 79.2 (77.1, 81.4) 5.5
5 (78.6, 82.6) 81.2 (79.2, 83.1) 5.8
6 (75.9, 80.2) 79.9 (77.9, 81.9) 6.6
7 (76.8, 81.0) 79.8 (77.8, 81.6) 7.9
8 (72.6, 77.7) 79.0 (76.9, 81.1) 5.5
9 (74.6, 79.5) 78.2 (76.2, 80.3) 9.8
10 (73.0, 77.5) 76.4 (74.1, 78.6) 6.4
CHD_Crude95CI CHD_AdjPrev CHD_Adj95CI CHECKUP_CrudePrev CHECKUP_Crude95CI
1 ( 6.7, 8.7) 6.1 ( 5.3, 6.9) 67.6 (63.1, 72.1)
2 ( 6.1, 8.0) 5.4 ( 4.8, 6.2) 71.6 (67.8, 75.2)
3 ( 8.0, 10.2) 7.1 ( 6.3, 8.0) 68.8 (64.8, 72.5)
4 ( 4.8, 6.2) 5.1 ( 4.5, 5.8) 64.1 (58.2, 69.7)
5 ( 5.1, 6.6) 5.2 ( 4.6, 5.9) 69.2 (66.1, 72.0)
6 ( 5.8, 7.5) 5.2 ( 4.6, 5.9) 69.3 (65.9, 72.5)
7 ( 6.9, 9.0) 5.7 ( 5.0, 6.4) 70.2 (66.0, 74.2)
8 ( 4.9, 6.3) 5.8 ( 5.2, 6.6) 63.3 (59.0, 67.6)
9 ( 8.5, 11.1) 6.0 ( 5.3, 6.8) 69.8 (65.6, 73.6)
10 ( 5.7, 7.3) 6.2 ( 5.4, 7.0) 66.8 (61.2, 72.2)
CHECKUP_AdjPrev CHECKUP_Adj95CI CHOLSCREEN_CrudePrev CHOLSCREEN_Crude95CI
1 65.5 (60.7, 70.2) 80.7 (77.5, 83.8)
2 68.8 (64.8, 72.7) 83.3 (80.7, 85.8)
3 65.8 (61.4, 69.8) 79.1 (76.1, 81.9)
4 63.4 (57.4, 69.0) 81.3 (78.0, 84.4)
5 68.0 (64.8, 71.0) 84.0 (81.7, 86.3)
6 66.9 (63.2, 70.4) 83.3 (80.9, 85.6)
7 66.4 (61.9, 70.7) 83.2 (80.4, 85.7)
8 63.8 (59.6, 67.9) 76.4 (73.2, 79.7)
9 63.1 (58.4, 67.4) 84.0 (81.5, 86.3)
10 66.4 (60.7, 71.8) 78.0 (74.3, 81.5)
CHOLSCREEN_AdjPrev CHOLSCREEN_Adj95CI COLON_SCREEN_CrudePrev
1 80.0 (76.5, 83.2) 63.2
2 80.9 (77.9, 83.7) 68.8
3 75.9 (72.7, 79.2) 59.9
4 80.7 (77.3, 84.0) 63.1
5 83.3 (80.8, 85.6) 67.8
6 82.1 (79.6, 84.7) 71.8
7 80.1 (76.9, 83.1) 69.2
8 79.3 (76.4, 82.3) 67.0
9 77.7 (74.3, 80.9) 67.4
10 78.1 (74.3, 81.6) 60.6
COLON_SCREEN_Crude95CI COLON_SCREEN_AdjPrev COLON_SCREEN_Adj95CI
1 (60.5, 65.7) 59.8 (56.7, 62.6)
2 (66.6, 71.1) 64.9 (62.5, 67.4)
3 (57.6, 62.2) 56.1 (53.7, 58.4)
4 (60.7, 65.7) 61.5 (59.1, 64.1)
5 (65.2, 70.2) 66.1 (63.3, 68.6)
6 (69.6, 74.0) 68.4 (66.0, 70.8)
7 (66.6, 71.5) 65.0 (62.4, 67.6)
8 (64.5, 69.3) 64.4 (61.9, 66.7)
9 (64.9, 69.8) 62.2 (59.5, 64.7)
10 (58.1, 63.2) 58.4 (55.8, 61.0)
COPD_CrudePrev COPD_Crude95CI COPD_AdjPrev COPD_Adj95CI COREM_CrudePrev
1 7.4 ( 6.1, 8.7) 6.4 ( 5.4, 7.5) 36.3
2 7.6 ( 6.3, 9.0) 6.3 ( 5.3, 7.5) 44.8
3 10.7 ( 9.1, 12.3) 8.9 ( 7.6, 10.2) 31.0
4 5.5 ( 4.7, 6.4) 5.2 ( 4.4, 6.1) 34.0
5 5.9 ( 4.9, 7.0) 5.4 ( 4.5, 6.4) 45.1
6 6.6 ( 5.4, 7.9) 5.6 ( 4.7, 6.7) 45.3
7 7.9 ( 6.5, 9.4) 6.2 ( 5.2, 7.4) 37.9
8 6.2 ( 5.2, 7.2) 6.4 ( 5.5, 7.5) 40.8
9 10.2 ( 8.5, 12.1) 7.3 ( 6.2, 8.6) 36.2
10 7.3 ( 6.2, 8.5) 7.0 ( 6.0, 8.3) 34.7
COREM_Crude95CI COREM_AdjPrev COREM_Adj95CI COREW_CrudePrev COREW_Crude95CI
1 (30.1, 42.5) 36.2 (30.1, 42.3) 29.0 (24.3, 34.1)
2 (38.3, 51.1) 44.7 (38.3, 50.8) 37.6 (31.7, 43.4)
3 (25.8, 36.0) 31.2 (26.1, 36.4) 22.1 (18.2, 26.0)
4 (27.9, 40.3) 34.8 (28.7, 40.8) 30.0 (24.9, 35.4)
5 (38.4, 51.0) 45.3 (38.8, 51.2) 36.2 (30.3, 41.5)
6 (39.1, 51.2) 45.5 (39.2, 51.3) 38.9 (33.3, 44.2)
7 (32.1, 43.7) 38.0 (32.2, 43.9) 34.8 (29.7, 39.9)
8 (34.6, 46.7) 41.4 (35.6, 47.0) 32.4 (27.4, 37.4)
9 (29.7, 42.9) 36.2 (29.9, 42.6) 29.6 (24.2, 35.4)
10 (28.8, 41.1) 35.0 (29.1, 41.5) 27.4 (22.8, 32.5)
COREW_AdjPrev COREW_Adj95CI CSMOKING_CrudePrev CSMOKING_Crude95CI
1 29.2 (24.3, 34.2) 14.5 (11.9, 17.4)
2 36.7 (31.1, 42.1) 16.6 (13.8, 19.6)
3 21.7 (18.0, 25.5) 20.9 (17.6, 24.1)
4 30.8 (25.7, 36.2) 14.9 (12.1, 17.8)
5 36.0 (30.3, 41.3) 13.8 (11.4, 16.3)
6 38.7 (33.1, 44.0) 14.0 (11.6, 16.6)
7 34.5 (29.5, 39.6) 15.5 (12.7, 18.5)
8 31.9 (27.3, 36.6) 14.5 (12.0, 17.4)
9 29.2 (24.0, 34.8) 18.4 (15.4, 21.9)
10 27.5 (23.0, 32.6) 18.8 (15.6, 22.3)
CSMOKING_AdjPrev CSMOKING_Adj95CI DENTAL_CrudePrev DENTAL_Crude95CI
1 15.7 (12.9, 18.8) 52.3 (48.7, 56.0)
2 17.2 (14.2, 20.2) 53.9 (50.8, 56.9)
3 21.9 (18.5, 25.3) 52.9 (49.8, 55.8)
4 14.9 (12.2, 17.9) 56.7 (53.5, 59.8)
5 14.0 (11.5, 16.6) 61.9 (58.9, 64.9)
6 15.1 (12.4, 17.8) 58.8 (55.8, 61.7)
7 16.5 (13.6, 19.9) 56.2 (53.2, 59.0)
8 16.2 (13.4, 19.5) 64.6 (61.5, 67.5)
9 20.4 (17.1, 24.0) 55.9 (52.4, 59.0)
10 19.3 (16.0, 22.8) 56.8 (53.8, 59.7)
DENTAL_AdjPrev DENTAL_Adj95CI DEPRESSION_CrudePrev DEPRESSION_Crude95CI
1 51.7 (47.8, 55.6) 17.9 (15.0, 21.0)
2 53.2 (50.0, 56.0) 17.9 (15.3, 20.5)
3 52.1 (49.1, 55.2) 20.3 (17.4, 23.3)
4 56.8 (53.5, 59.9) 18.3 (14.9, 21.9)
5 61.7 (58.7, 64.7) 18.7 (16.7, 20.9)
6 58.0 (54.8, 61.0) 21.8 (19.2, 24.4)
7 55.3 (52.2, 58.2) 19.4 (16.6, 22.7)
8 63.8 (60.8, 66.5) 20.5 (17.6, 23.7)
9 54.2 (51.3, 57.2) 18.8 (16.0, 22.0)
10 56.7 (53.7, 59.6) 19.6 (16.1, 23.4)
DEPRESSION_AdjPrev DEPRESSION_Adj95CI DIABETES_CrudePrev DIABETES_Crude95CI
1 18.4 (15.4, 21.6) 14.3 (12.4, 16.3)
2 18.5 (15.8, 21.3) 12.1 (10.6, 13.8)
3 21.1 (18.1, 24.1) 15.4 (13.5, 17.4)
4 18.4 (15.0, 22.1) 10.1 ( 8.6, 11.6)
5 18.9 (16.9, 21.1) 10.4 ( 9.2, 11.5)
6 22.4 (19.7, 25.0) 10.5 ( 9.2, 11.9)
7 20.4 (17.5, 23.8) 12.2 (10.6, 14.0)
8 19.9 (17.2, 23.0) 9.0 ( 7.9, 10.4)
9 20.6 (17.6, 24.0) 14.0 (12.2, 16.0)
10 19.6 (16.1, 23.4) 11.3 ( 9.7, 13.1)
DIABETES_AdjPrev DIABETES_Adj95CI GHLTH_CrudePrev GHLTH_Crude95CI
1 13.0 (11.3, 14.8) 23.2 (20.0, 26.8)
2 10.1 ( 8.9, 11.5) 18.4 (15.7, 21.2)
3 12.8 (11.1, 14.4) 24.2 (21.2, 27.3)
4 9.6 ( 8.2, 11.0) 16.4 (14.1, 18.8)
5 9.5 ( 8.5, 10.6) 15.4 (13.2, 17.6)
6 9.1 ( 7.9, 10.3) 17.5 (15.0, 20.0)
7 9.7 ( 8.4, 11.2) 18.7 (15.9, 21.4)
8 9.5 ( 8.3, 11.0) 14.8 (12.8, 17.2)
9 9.8 ( 8.6, 11.2) 21.5 (18.7, 24.7)
10 10.9 ( 9.3, 12.7) 20.0 (17.2, 22.9)
GHLTH_AdjPrev GHLTH_Adj95CI HIGHCHOL_CrudePrev HIGHCHOL_Crude95CI
1 22.4 (19.3, 25.9) 36.3 (32.6, 40.1)
2 17.2 (14.6, 19.8) 37.7 (34.4, 41.0)
3 22.5 (19.8, 25.3) 36.2 (32.8, 39.4)
4 16.1 (13.8, 18.5) 30.8 (26.6, 35.4)
5 14.8 (12.8, 17.0) 34.5 (32.2, 36.9)
6 16.6 (14.2, 18.9) 35.2 (32.5, 37.9)
7 17.0 (14.5, 19.5) 38.3 (34.9, 42.0)
8 15.5 (13.4, 17.8) 29.4 (26.6, 32.4)
9 18.6 (16.2, 21.5) 36.1 (32.4, 39.7)
10 19.8 (17.1, 22.7) 34.3 (30.0, 38.8)
HIGHCHOL_AdjPrev HIGHCHOL_Adj95CI KIDNEY_CrudePrev KIDNEY_Crude95CI
1 31.2 (27.4, 35.1) 4.2 ( 3.8, 4.6)
2 31.3 (27.9, 34.6) 3.7 ( 3.3, 4.1)
3 28.8 (25.6, 32.0) 4.6 ( 4.2, 5.1)
4 27.5 (23.3, 32.2) 3.2 ( 2.9, 3.5)
5 30.6 (28.2, 33.1) 3.3 ( 3.0, 3.6)
6 29.7 (27.0, 32.5) 3.7 ( 3.3, 4.1)
7 30.6 (27.3, 34.4) 4.0 ( 3.6, 4.4)
8 27.5 (24.4, 30.8) 3.1 ( 2.8, 3.4)
9 25.8 (22.6, 29.0) 4.8 ( 4.4, 5.3)
10 30.8 (26.3, 35.5) 3.5 ( 3.2, 3.8)
KIDNEY_AdjPrev KIDNEY_Adj95CI LPA_CrudePrev LPA_Crude95CI LPA_AdjPrev
1 3.6 ( 3.2, 3.9) 31.2 (27.0, 36.1) 30.5
2 3.1 ( 2.8, 3.3) 25.4 (21.7, 29.3) 24.3
3 3.8 ( 3.5, 4.2) 27.1 (23.2, 31.0) 25.9
4 3.1 ( 2.8, 3.4) 23.3 (19.2, 27.7) 23.1
5 3.0 ( 2.7, 3.3) 22.4 (19.4, 25.5) 22.0
6 3.1 ( 2.8, 3.4) 24.2 (20.8, 27.6) 23.5
7 3.1 ( 2.8, 3.4) 26.7 (22.6, 31.0) 25.2
8 3.3 ( 3.0, 3.6) 17.7 (15.0, 20.8) 18.5
9 3.3 ( 3.0, 3.7) 28.1 (23.9, 32.7) 25.5
10 3.4 ( 3.1, 3.7) 25.7 (21.4, 30.4) 25.6
LPA_Adj95CI MAMMOUSE_CrudePrev MAMMOUSE_Crude95CI MAMMOUSE_AdjPrev
1 (26.1, 35.2) 65.2 (61.4, 68.8) 66.2
2 (20.7, 27.9) 66.1 (62.4, 69.8) 67.1
3 (22.2, 29.6) 57.4 (53.8, 60.9) 57.3
4 (18.8, 27.5) 67.8 (64.1, 71.4) 67.1
5 (19.0, 25.1) 69.0 (65.5, 72.3) 69.1
6 (20.1, 26.8) 70.4 (67.1, 73.6) 70.9
7 (21.2, 29.5) 69.4 (65.9, 72.7) 69.9
8 (15.8, 21.7) 62.5 (59.2, 65.8) 61.8
9 (21.8, 29.7) 61.6 (57.6, 65.3) 62.2
10 (21.2, 30.2) 62.3 (58.8, 66.0) 62.3
MAMMOUSE_Adj95CI MHLTH_CrudePrev MHLTH_Crude95CI MHLTH_AdjPrev MHLTH_Adj95CI
1 (62.3, 69.9) 15.8 (13.6, 18.1) 16.4 (14.2, 18.8)
2 (63.5, 70.5) 15.0 (13.1, 16.9) 16.0 (14.1, 18.0)
3 (53.6, 60.7) 18.7 (16.4, 20.8) 20.1 (17.7, 22.4)
4 (63.3, 71.0) 15.3 (13.2, 17.4) 15.6 (13.5, 17.7)
5 (65.7, 72.5) 16.1 (14.5, 17.8) 16.5 (14.8, 18.3)
6 (67.5, 73.9) 16.2 (14.4, 18.1) 17.0 (15.0, 19.0)
7 (66.3, 73.2) 14.2 (12.4, 16.2) 15.5 (13.6, 17.6)
8 (58.4, 65.2) 18.2 (15.9, 20.8) 17.4 (15.4, 19.7)
9 (58.3, 65.8) 16.1 (14.2, 18.3) 19.0 (16.7, 21.5)
10 (58.6, 65.9) 17.7 (15.3, 20.1) 17.8 (15.3, 20.2)
OBESITY_CrudePrev OBESITY_Crude95CI OBESITY_AdjPrev OBESITY_Adj95CI
1 39.1 (33.6, 44.9) 40.8 (35.1, 46.7)
2 37.2 (32.6, 41.9) 37.5 (32.9, 42.2)
3 38.0 (33.0, 42.6) 38.2 (33.3, 42.9)
4 31.1 (24.2, 38.6) 31.1 (24.3, 38.5)
5 30.8 (27.4, 34.3) 30.9 (27.6, 34.4)
6 31.8 (28.0, 35.7) 32.9 (28.9, 36.9)
7 33.5 (28.8, 38.6) 34.3 (29.4, 39.5)
8 24.6 (20.8, 28.9) 26.4 (22.5, 30.9)
9 35.9 (31.0, 41.0) 36.6 (31.7, 41.7)
10 35.9 (29.2, 43.1) 36.3 (29.6, 43.6)
PHLTH_CrudePrev PHLTH_Crude95CI PHLTH_AdjPrev PHLTH_Adj95CI SLEEP_CrudePrev
1 14.1 (12.1, 16.1) 13.5 (11.6, 15.5) 35.1
2 12.9 (11.2, 14.8) 12.0 (10.4, 13.6) 35.7
3 16.5 (14.5, 18.5) 15.3 (13.4, 17.1) 37.7
4 11.2 ( 9.7, 12.8) 11.0 ( 9.5, 12.4) 33.4
5 10.9 ( 9.5, 12.3) 10.5 ( 9.2, 11.9) 31.7
6 12.1 (10.5, 13.7) 11.4 ( 9.9, 13.0) 32.5
7 13.0 (11.2, 14.8) 11.8 (10.1, 13.4) 33.1
8 11.0 ( 9.6, 12.6) 11.6 (10.2, 13.2) 28.8
9 15.3 (13.3, 17.5) 13.2 (11.5, 15.0) 34.5
10 13.2 (11.5, 15.1) 13.1 (11.4, 14.9) 34.1
SLEEP_Crude95CI SLEEP_AdjPrev SLEEP_Adj95CI STROKE_CrudePrev
1 (33.9, 36.3) 36.6 (35.3, 37.9) 4.0
2 (34.6, 36.8) 37.0 (35.8, 38.0) 3.6
3 (36.5, 38.7) 39.0 (37.8, 40.0) 5.0
4 (32.1, 34.6) 33.6 (32.3, 34.9) 2.9
5 (30.6, 32.7) 32.2 (31.1, 33.3) 2.9
6 (31.4, 33.6) 34.0 (32.7, 35.2) 3.4
7 (31.8, 34.2) 34.7 (33.4, 35.9) 3.9
8 (27.6, 29.9) 29.9 (28.8, 31.0) 3.0
9 (33.3, 35.6) 37.1 (35.9, 38.1) 4.9
10 (33.1, 35.2) 34.6 (33.5, 35.7) 3.4
STROKE_Crude95CI STROKE_AdjPrev STROKE_Adj95CI TEETHLOST_CrudePrev
1 ( 3.5, 4.5) 3.3 ( 2.9, 3.7) 13.7
2 ( 3.2, 4.1) 2.9 ( 2.6, 3.3) 12.8
3 ( 4.4, 5.6) 4.1 ( 3.7, 4.5) 17.4
4 ( 2.6, 3.2) 2.7 ( 2.4, 3.0) 10.7
5 ( 2.6, 3.3) 2.7 ( 2.4, 3.1) 10.3
6 ( 3.0, 3.9) 2.8 ( 2.5, 3.2) 10.6
7 ( 3.4, 4.3) 2.9 ( 2.6, 3.3) 12.9
8 ( 2.7, 3.3) 3.2 ( 2.8, 3.5) 11.5
9 ( 4.3, 5.6) 3.3 ( 2.9, 3.7) 13.4
10 ( 3.1, 3.8) 3.3 ( 3.0, 3.7) 13.3
TEETHLOST_Crude95CI TEETHLOST_AdjPrev TEETHLOST_Adj95CI HEARING_CrudePrev
1 ( 9.9, 17.8) 14.6 (10.7, 18.9) 9.1
2 ( 8.9, 17.5) 13.2 ( 9.3, 18.3) 8.2
3 (13.4, 21.9) 18.2 (13.9, 22.9) 10.5
4 ( 7.7, 13.6) 11.5 ( 8.4, 14.8) 6.7
5 ( 6.7, 14.7) 10.8 ( 7.1, 15.6) 6.6
6 ( 7.1, 14.8) 11.2 ( 7.4, 15.8) 7.7
7 ( 9.2, 17.2) 13.5 ( 9.5, 18.2) 8.7
8 ( 8.2, 15.1) 12.0 ( 8.6, 15.7) 6.8
9 ( 9.4, 18.2) 14.0 ( 9.7, 19.2) 11.3
10 (10.0, 17.1) 14.3 (10.5, 18.6) 7.8
HEARING_Crude95CI HEARING_AdjPrev HEARING_Adj95CI VISION_CrudePrev
1 ( 8.1, 10.2) 7.4 ( 6.6, 8.3) 8.2
2 ( 7.2, 9.2) 6.7 ( 6.0, 7.5) 5.4
3 ( 9.4, 11.7) 8.9 ( 7.9, 9.8) 8.6
4 ( 5.9, 7.5) 6.4 ( 5.7, 7.1) 5.2
5 ( 5.8, 7.3) 6.1 ( 5.4, 6.8) 4.7
6 ( 6.8, 8.6) 6.4 ( 5.7, 7.1) 5.5
7 ( 7.7, 9.8) 6.7 ( 5.9, 7.5) 6.0
8 ( 6.0, 7.6) 7.2 ( 6.4, 8.0) 5.1
9 (10.0, 12.7) 7.7 ( 6.9, 8.7) 7.1
10 ( 6.9, 8.8) 7.6 ( 6.7, 8.5) 6.6
VISION_Crude95CI VISION_AdjPrev VISION_Adj95CI COGNITION_CrudePrev
1 ( 7.1, 9.5) 7.8 ( 6.7, 9.0) 16.3
2 ( 4.7, 6.3) 5.1 ( 4.4, 5.9) 13.8
3 ( 7.5, 9.7) 8.0 ( 7.0, 9.0) 18.4
4 ( 4.5, 5.9) 5.1 ( 4.5, 5.8) 13.5
5 ( 4.1, 5.3) 4.5 ( 3.9, 5.2) 13.2
6 ( 4.7, 6.3) 5.1 ( 4.4, 5.9) 13.2
7 ( 5.1, 6.9) 5.4 ( 4.7, 6.2) 13.8
8 ( 4.4, 5.8) 5.3 ( 4.6, 6.0) 14.8
9 ( 6.1, 8.1) 6.1 ( 5.3, 7.0) 15.1
10 ( 5.8, 7.5) 6.5 ( 5.7, 7.4) 16.6
COGNITION_Crude95CI COGNITION_AdjPrev COGNITION_Adj95CI MOBILITY_CrudePrev
1 (13.9, 19.0) 16.5 (14.1, 19.2) 16.9
2 (11.8, 15.8) 14.4 (12.3, 16.5) 14.9
3 (16.0, 20.7) 19.2 (16.7, 21.6) 20.1
4 (11.5, 15.5) 13.7 (11.6, 15.7) 12.2
5 (11.5, 15.1) 13.4 (11.6, 15.4) 12.7
6 (11.4, 15.2) 13.5 (11.6, 15.6) 14.1
7 (11.9, 16.0) 14.5 (12.5, 16.7) 15.7
8 (12.5, 17.3) 14.2 (12.3, 16.4) 11.0
9 (13.0, 17.5) 16.7 (14.6, 19.3) 20.7
10 (14.2, 19.0) 16.6 (14.2, 19.0) 14.9
MOBILITY_Crude95CI MOBILITY_AdjPrev MOBILITY_Adj95CI SELFCARE_CrudePrev
1 (14.6, 19.3) 15.0 (12.8, 17.2) 5.5
2 (12.8, 17.2) 12.6 (10.9, 14.6) 4.0
3 (17.7, 22.5) 17.1 (15.0, 19.2) 6.3
4 (10.5, 14.0) 11.7 (10.0, 13.4) 3.6
5 (11.0, 14.5) 11.8 (10.2, 13.5) 3.4
6 (12.1, 16.1) 12.2 (10.5, 14.0) 3.9
7 (13.5, 18.1) 12.6 (10.8, 14.5) 4.4
8 ( 9.6, 12.7) 11.8 (10.3, 13.5) 3.5
9 (17.9, 23.8) 15.0 (13.0, 17.2) 5.5
10 (12.8, 17.0) 14.5 (12.5, 16.7) 4.6
SELFCARE_Crude95CI SELFCARE_AdjPrev SELFCARE_Adj95CI INDEPLIVE_CrudePrev
1 ( 4.7, 6.3) 5.2 ( 4.5, 6.0) 9.6
2 ( 3.5, 4.6) 3.7 ( 3.2, 4.2) 7.8
3 ( 5.5, 7.2) 5.7 ( 5.0, 6.4) 11.7
4 ( 3.2, 4.1) 3.5 ( 3.1, 4.0) 6.9
5 ( 2.9, 4.0) 3.3 ( 2.8, 3.8) 6.8
6 ( 3.4, 4.5) 3.7 ( 3.1, 4.2) 7.6
7 ( 3.8, 5.1) 3.9 ( 3.4, 4.4) 8.0
8 ( 3.1, 4.0) 3.9 ( 3.4, 4.4) 7.9
9 ( 4.8, 6.4) 4.5 ( 3.9, 5.1) 9.9
10 ( 4.1, 5.3) 4.6 ( 4.1, 5.2) 9.1
INDEPLIVE_Crude95CI INDEPLIVE_AdjPrev INDEPLIVE_Adj95CI DISABILITY_CrudePrev
1 ( 8.3, 11.1) 9.2 ( 7.9, 10.6) 33.3
2 ( 6.6, 8.9) 7.5 ( 6.4, 8.7) 31.0
3 (10.2, 13.3) 11.4 (10.0, 12.9) 37.8
4 ( 6.0, 7.9) 6.9 ( 6.0, 7.9) 27.6
5 ( 5.9, 7.9) 6.7 ( 5.8, 7.8) 28.5
6 ( 6.6, 8.8) 7.4 ( 6.3, 8.4) 30.1
7 ( 6.9, 9.2) 7.6 ( 6.6, 8.8) 31.8
8 ( 6.8, 9.1) 8.0 ( 6.9, 9.2) 27.1
9 ( 8.5, 11.4) 9.2 ( 8.0, 10.6) 38.7
10 ( 7.9, 10.4) 9.0 ( 7.9, 10.3) 33.4
DISABILITY_Crude95CI DISABILITY_AdjPrev DISABILITY_Adj95CI
1 (28.9, 37.6) 30.9 (26.7, 35.1)
2 (27.0, 34.7) 28.8 (25.1, 32.4)
3 (33.8, 41.6) 35.5 (31.7, 39.4)
4 (23.7, 31.6) 27.1 (23.1, 31.1)
5 (25.5, 31.5) 27.6 (24.7, 30.6)
6 (26.6, 33.6) 27.9 (24.7, 31.3)
7 (28.2, 35.9) 28.7 (25.2, 32.6)
8 (23.7, 30.9) 27.2 (24.1, 30.9)
9 (34.6, 43.1) 33.9 (30.0, 37.8)
10 (29.1, 38.0) 32.9 (28.6, 37.5)
Geolocation geometry
1 POINT (-113.910905 32.7739424) MULTIPOLYGON (((-114.8141 3...
2 POINT (-111.3663396 32.9185209) MULTIPOLYGON (((-111.2669 3...
3 POINT (-110.3210248 35.3907852) MULTIPOLYGON (((-110.0007 3...
4 POINT (-109.2423231 33.2388723) MULTIPOLYGON (((-109.4958 3...
5 POINT (-112.4989296 33.3451756) MULTIPOLYGON (((-111.8931 3...
6 POINT (-111.7836574 32.128038) MULTIPOLYGON (((-111.0388 3...
7 POINT (-109.7751627 31.8401287) MULTIPOLYGON (((-110.4523 3...
8 POINT (-111.7737277 35.8296919) MULTIPOLYGON (((-112.6604 3...
9 POINT (-110.8118696 33.7896177) MULTIPOLYGON (((-111.7207 3...
10 POINT (-109.8783103 32.9318277) MULTIPOLYGON (((-110.4494 3...Table to Points
- Tables with longitude and latitude data can be convered into
sfobjects. - Addresses needed to be converted to longitude and latitude data before conversion to
sfobjects. This process is called geocoding. st_as_sf(df, coords = c("lon", "lat"), crs =crs)
Table to Points
Simple feature collection with 85395 features and 4 fields
Geometry type: POINT
Dimension: XY
Bounding box: xmin: -175.86 ymin: 17.90163 xmax: -65.30188 ymax: 71.29667
Geodetic CRS: WGS 84
# A tibble: 85,395 × 5
STATEFP COUNTYFP TRACTCE POPULATION geometry
* <chr> <chr> <chr> <dbl> <POINT [°]>
1 01 001 020100 1775 (-86.4866 32.47682)
2 01 001 020200 2055 (-86.47259 32.4719)
3 01 001 020300 3216 (-86.45925 32.47458)
4 01 001 020400 4246 (-86.44299 32.46866)
5 01 001 020501 4322 (-86.42442 32.45153)
6 01 001 020502 3284 (-86.41773 32.46832)
7 01 001 020503 3616 (-86.42498 32.47547)
8 01 001 020600 3729 (-86.47658 32.44332)
9 01 001 020700 3409 (-86.44874 32.44379)
10 01 001 020801 3143 (-86.52377 32.44077)
# ℹ 85,385 more rowsSpatial Joins
- Joins on a spatial relationship, including
st_intersects()st_contains()st_is_within_distance()
- Takes geometry from first
sfobject st_join(sf1,sf2, join =One of above)
Spatial Joins
Simple feature collection with 157 features and 60 fields
Geometry type: POINT
Dimension: XY
Bounding box: xmin: -114.6334 ymin: 31.341 xmax: -109.04 ymax: 36.91705
Geodetic CRS: WGS 84
First 10 features:
OBJECTID OBJECTID_1 RUN_DATE source BUREAU FACID
1 1 2822 2024-05-06 ASPEN MED AZTH00002
2 2 2823 2024-05-06 ASPEN MED AZTH00003
3 3 2824 2024-05-06 ASPEN MED AZTH00004
4 4 3774 2024-05-06 ASPEN MED BH7347
5 5 4893 2024-05-06 ASPEN MED MED0198
6 6 4894 2024-05-06 ASPEN MED MED0201
7 7 4895 2024-05-06 ASPEN MED MED0203
8 8 4896 2024-05-06 ASPEN MED MED0204
9 9 4901 2024-05-06 ASPEN MED MED0209
10 10 4902 2024-05-06 ASPEN MED MED0211
FACILITY_N LICENSE_NU LICENSE_EF
1 BANNER- UNIVERSITY MEDICAL CENTER PHOENIX <NA> <NA>
2 BANNER UNIVERSITY MEDICAL CTR AT THE AZ HEALTH SC <NA> <NA>
3 MAYO CLINIC HOSPITAL <NA> <NA>
4 VIA LINDA BEHAVIORAL HOSPITAL SH11520 2023-03-01
5 CANYON VISTA MEDICAL CENTER H7130 2020-02-18
6 FLAGSTAFF MEDICAL CENTER H0169 2020-12-01
7 PAGE HOSPITAL H0086 2022-01-06
8 BANNER PAYSON MEDICAL CENTER H7250 2021-10-15
9 ABRAZO ARROWHEAD CAMPUS H0175 2021-12-01
10 BANNER BEHAVIORAL HEALTH HOSPITAL SH0147 2021-02-01
LICENSE_EX MEDICARE_I MEDICARE_1 MEDICARE_2 TELEPHONE FACILITY_T
1 <NA> 039802 <NA> <NA> (602)239-2716 NA
2 <NA> 039800 <NA> <NA> (520)694-0111 NA
3 <NA> 039801 <NA> <NA> (480)342-1900 NA
4 2025-02-28 034040 <NA> <NA> (480)476-7000 12
5 2024-12-27 030043 <NA> <NA> (520)263-2220 11
6 2024-11-30 030023 <NA> <NA> (928)779-3366 11
7 2024-12-28 031304 <NA> <NA> (928)645-2424 14
8 2024-12-24 031318 <NA> <NA> (928)471-3222 14
9 2024-12-27 030094 <NA> <NA> (623)561-1000 11
10 2024-12-24 034004 <NA> <NA> (480)448-7500 12
TYPE SUBTYPE CATEGORY ICON_CATEG
1 HOSPITAL TRANSPLANT HOSPITAL HOSPITAL
2 HOSPITAL TRANSPLANT HOSPITAL HOSPITAL
3 HOSPITAL TRANSPLANT HOSPITAL HOSPITAL
4 HOSPITAL PSYCHIATRIC HOSPITAL HOSPITAL
5 HOSPITAL SHORT TERM HOSPITAL HOSPITAL
6 HOSPITAL SHORT TERM HOSPITAL HOSPITAL
7 HOSPITAL CRITICAL ACCESS HOSPITALS HOSPITAL HOSPITAL
8 HOSPITAL CRITICAL ACCESS HOSPITALS HOSPITAL HOSPITAL
9 HOSPITAL SHORT TERM HOSPITAL HOSPITAL
10 HOSPITAL PSYCHIATRIC HOSPITAL HOSPITAL
MEDICARE_T LICENSE_TY LICENSE_SU
1 HOSPITAL - TRANSPLANT HOSPITAL FEDERAL ONLY <NA>
2 HOSPITAL - TRANSPLANT HOSPITAL FEDERAL ONLY <NA>
3 HOSPITAL - TRANSPLANT HOSPITAL FEDERAL ONLY <NA>
4 HOSPITAL - PSYCHIATRIC HOSPITAL HOSPITAL - SPECIAL
5 HOSPITAL - SHORT TERM HOSPITAL HOSPITAL - GENERAL
6 HOSPITAL - SHORT TERM HOSPITAL HOSPITAL - GENERAL
7 HOSPITAL - CRITICAL ACCESS HOSPITALS HOSPITAL HOSPITAL - GENERAL
8 HOSPITAL - CRITICAL ACCESS HOSPITALS HOSPITAL HOSPITAL - GENERAL
9 HOSPITAL - SHORT TERM HOSPITAL HOSPITAL - GENERAL
10 HOSPITAL - PSYCHIATRIC HOSPITAL HOSPITAL - SPECIAL
CAPACITY ADDRESS CITY ZIP COUNTY OPERATION_
1 0 1441 N 12TH PHOENIX 85006 MARICOPA ACTIVE
2 0 1501 NORTH CAMPBELL AVENUE TUCSON 85724 PIMA ACTIVE
3 0 5777 EAST MAYO BOULEVARD PHOENIX 85054 MARICOPA ACTIVE
4 120 9160 EAST HORSESHOE RD SCOTTSDALE 85258 MARICOPA ACTIVE
5 100 5700 EAST HIGHWAY 90 SIERRA VISTA 85635 COCHISE ACTIVE
6 268 1200 NORTH BEAVER STREET FLAGSTAFF 86001 COCONINO ACTIVE
7 25 501 NORTH NAVAJO DRIVE PAGE 86040 COCONINO ACTIVE
8 25 807 SOUTH PONDEROSA DRIVE PAYSON 85541 GILA ACTIVE
9 229 18701 NORTH 67TH AVENUE GLENDALE 85308 MARICOPA ACTIVE
10 156 7575 EAST EARLL DRIVE SCOTTSDALE 85251 MARICOPA ACTIVE
X_Is_Publi HOSPITAL_G KeyID ADHSCODE N_AddressT N_LocatorT
1 Yes <NA> AZTH00002 A AS0 CENTRUS-TOMTOM
2 Yes <NA> AZTH00003 A AS0 CENTRUS-TOMTOM
3 Yes <NA> AZTH00004 A AS0 CENTRUS-TOMTOM
4 Yes <NA> BH7347 A AS0 CENTRUS-TOMTOM
5 Yes <NA> MED0198 A AS0 CENTRUS-TOMTOM
6 Yes <NA> MED0201 A AS0 CENTRUS-TOMTOM
7 Yes <NA> MED0203 A AS0 CENTRUS-TOMTOM
8 Yes <NA> MED0204 A AS0 CENTRUS-TOMTOM
9 Yes <NA> MED0209 A AS0 CENTRUS-TOMTOM
10 Yes <NA> MED0211 A AS0 CENTRUS-TOMTOM
N_ADDRESS N_ADDR2 N_CITY N_COUNTY
1 1441 N 12TH ST <NA> PHOENIX MARICOPA COUNTY
2 1501 N CAMPBELL AVE <NA> TUCSON PIMA COUNTY
3 5777 E MAYO BLVD <NA> PHOENIX MARICOPA COUNTY
4 9160 E HORSESHOE RD <NA> SCOTTSDALE MARICOPA COUNTY
5 5700 E HIGHWAY 90 <NA> SIERRA VISTA COCHISE COUNTY
6 1200 N BEAVER ST <NA> FLAGSTAFF COCONINO COUNTY
7 501 N NAVAJO DR <NA> PAGE COCONINO COUNTY
8 807 S PONDEROSA ST <NA> PAYSON GILA COUNTY
9 18701 N 67TH AVE <NA> GLENDALE MARICOPA COUNTY
10 7575 E EARLL DR <NA> SCOTTSDALE MARICOPA COUNTY
N_FULLADDR N_LAT N_LON N_STATE
1 1441 N 12TH ST , PHOENIX, AZ 85006-2837 33.46410 -112.0562 AZ
2 1501 N CAMPBELL AVE , TUCSON, AZ 85724-0001 32.24080 -110.9443 AZ
3 5777 E MAYO BLVD , PHOENIX, AZ 85054-4502 33.66316 -111.9557 AZ
4 9160 E HORSESHOE RD , SCOTTSDALE, AZ 85258-4666 33.56670 -111.8840 AZ
5 5700 E HIGHWAY 90 , SIERRA VISTA, AZ 85635-9110 31.55449 -110.2319 AZ
6 1200 N BEAVER ST , FLAGSTAFF, AZ 86001-3118 35.20949 -111.6447 AZ
7 501 N NAVAJO DR , PAGE, AZ 86040-0959 36.91705 -111.4634 AZ
8 807 S PONDEROSA ST , PAYSON, AZ 85541-5542 34.23130 -111.3212 AZ
9 18701 N 67TH AVE , GLENDALE, AZ 85308-7100 33.65504 -112.2027 AZ
10 7575 E EARLL DR , SCOTTSDALE, AZ 85251-6915 33.48385 -111.9183 AZ
N_ZIP N_ZIP4 N_BLOCK RESERVATIO RESERV_ID R_METHOD
1 85006 2837 4.013113e+13 <NA> <NA> <NA>
2 85724 1 4.019002e+13 <NA> <NA> <NA>
3 85054 4502 4.013615e+13 <NA> <NA> <NA>
4 85258 4666 4.013941e+13 Salt River Reservation 3340F Coordinates
5 85635 9110 4.003002e+13 <NA> <NA> <NA>
6 86001 3118 4.005000e+13 <NA> <NA> <NA>
7 86040 959 4.005002e+13 <NA> <NA> <NA>
8 85541 5542 4.007001e+13 <NA> <NA> <NA>
9 85308 7100 4.013616e+13 <NA> <NA> <NA>
10 85251 6915 4.013218e+13 <NA> <NA> <NA>
oPCA oPCA_ID P_Method P_Reliable
1 CENTRAL CITY VILLAGE 39 Coordinates <NA>
2 TUCSON CENTRAL 107 Coordinates <NA>
3 DESERT VIEW VILLAGE 30 Coordinates <NA>
4 SALT RIVER PIMA-MARICOPA INDIAN COMMUNITY 62 Coordinates <NA>
5 SIERRA VISTA 123 Coordinates <NA>
6 FLAGSTAFF 10 Coordinates <NA>
7 PAGE 7 Coordinates <NA>
8 PAYSON 23 Coordinates <NA>
9 GLENDALE NORTH 56 Coordinates <NA>
10 SCOTTSDALE SOUTH 45 Coordinates <NA>
SOURCE_VW CASELOAD CSA_ID
1 Tableau.vw_licensing_facilities <NA> 47
2 Tableau.vw_licensing_facilities <NA> 121
3 Tableau.vw_licensing_facilities <NA> 37
4 Tableau.vw_licensing_facilities <NA> 72
5 Tableau.vw_licensing_facilities <NA> 135
6 Tableau.vw_licensing_facilities <NA> 11
7 Tableau.vw_licensing_facilities <NA> 8
8 Tableau.vw_licensing_facilities <NA> 30
9 Tableau.vw_licensing_facilities <NA> 65
10 Tableau.vw_licensing_facilities <NA> 54
CSA_NAME AREA_SQMILE POP2020 Shape__Area
1 Phoenix - Central City 26.06187 62645 726562975
2 Tucson - North 31.92997 124515 890156111
3 Phoenix - Desert View 62.87998 61105 1752992698
4 Salt River Pima-Maricopa Indian Community 84.99103 6334 2369413211
5 Sierra Vista 708.44630 56911 19750344116
6 Flagstaff 4579.81300 88600 127677843904
7 Page 3160.98200 9375 88123101547
8 Payson 2106.17700 28134 58716828639
9 Glendale - North 26.11177 90431 727954288
10 Scottsdale - South 16.24433 84604 452865825
Shape__Length geometry
1 147316.9 POINT (-112.0562 33.46411)
2 174100.1 POINT (-110.9443 32.24081)
3 222767.7 POINT (-111.9557 33.66317)
4 259861.9 POINT (-111.884 33.56671)
5 905907.7 POINT (-110.2319 31.5545)
6 3525132.7 POINT (-111.6447 35.20949)
7 2193590.5 POINT (-111.4634 36.91705)
8 1943665.6 POINT (-111.3212 34.2313)
9 150309.6 POINT (-112.2027 33.65505)
10 108575.4 POINT (-111.9183 33.48386)Spatial Join + Summarize
st_join(AZ_hosp, AZ_statAreas) %>%
group_by(CSA_ID) %>%
summarize(count = n(), POP2020 = first(POP2020)) %>%
mutate(countPerHunThoP = 100000 * count / POP2020)Simple feature collection with 81 features and 4 fields
Geometry type: GEOMETRY
Dimension: XY
Bounding box: xmin: -114.6334 ymin: 31.341 xmax: -109.04 ymax: 36.91705
Geodetic CRS: WGS 84
# A tibble: 81 × 5
CSA_ID count POP2020 geometry countPerHunThoP
* <int> <int> <int> <GEOMETRY [°]> <dbl>
1 4 2 54625 MULTIPOINT ((-114.0359 35.21854), (-113… 3.66
2 5 2 41406 MULTIPOINT ((-114.5982 35.10075), (-114… 4.83
3 6 1 25119 POINT (-114.5978 35.00245) 3.98
4 7 1 60701 POINT (-114.3387 34.47989) 1.65
5 8 1 9375 POINT (-111.4634 36.91705) 10.7
6 9 2 90302 MULTIPOINT ((-109.59 36.14001), (-109.5… 2.21
7 11 3 88600 MULTIPOINT ((-111.6062 35.20678), (-111… 3.39
8 12 2 10321 MULTIPOINT ((-110.42 35.80001), (-111.2… 19.4
9 15 1 16962 POINT (-110.6919 35.03552) 5.90
10 17 2 29859 MULTIPOINT ((-109.9751 34.16484), (-110… 6.70
# ℹ 71 more rowsBuffers
- Gives as the crow flies distance around a point, line or polygon
- Benefits greatly from a good projection
- Can help us get at questions of access
st_buffer(sf1,distance)
Buffers
Distance
- Allows us to tell how far two locations are
- Can help us get at questions of access
- By default, will generate distances between every element of one sf to another
st_distance(sf1,sf2, by_element =TRUE/FALSE)
Distance Matrix
read_csv("data/US/COP2020/COP2020_tract.txt") %>%
filter(STATEFP == "04") %>%
st_as_sf(coords = c("LONGITUDE", "LATITUDE"), crs = 4326) %>%
st_distance(., AZ_hosp) %>%
head()Units: [m]
[,1] [,2] [,3] [,4] [,5] [,6] [,7] [,8]
[1,] 418834.8 513413.9 394927.7 401342.2 580851.5 237198.5 144967.4 312153.8
[2,] 442157.8 524284.0 418543.8 423948.9 585561.1 269397.1 188378.8 333760.8
[3,] 374473.8 429418.4 352467.7 355090.9 481961.5 240075.2 247279.7 266714.7
[4,] 411224.3 484801.1 388013.6 392518.2 543509.6 249879.8 203963.1 302232.5
[5,] 375571.1 454554.7 352226.4 357013.0 517079.5 213458.0 185629.9 266685.4
[6,] 378943.7 455596.3 355691.1 360286.5 517005.9 218844.0 191983.3 269977.8
[,9] [,10] [,11] [,12] [,13] [,14] [,15] [,16]
[1,] 407064.0 411000.9 418508.9 415022.4 418765.5 401926.9 419477.3 419011.2
[2,] 432334.1 433450.8 440203.7 441386.5 442107.7 426685.4 442646.6 441275.4
[3,] 370145.0 363863.7 368538.6 381909.9 374474.5 363319.9 374554.8 370965.1
[4,] 403209.3 401830.4 407926.3 413249.6 411191.1 397124.0 411578.4 409455.4
[5,] 367076.2 366395.4 372733.7 376925.1 375532.7 361087.8 375967.3 374094.9
[6,] 370812.4 369624.9 375816.6 380840.0 378908.8 364742.6 379311.5 377278.7
[,17] [,18] [,19] [,20] [,21] [,22] [,23] [,24]
[1,] 410801.4 415688.1 462340.2 479213.2 414507.0 515745.6 511192.3 513245.8
[2,] 433302.9 441213.3 503204.4 517404.3 455078.6 527002.8 521601.9 524119.3
[3,] 363849.6 379399.9 500799.9 498919.0 452229.4 432706.6 426063.2 429260.9
[4,] 401726.5 412271.1 496611.6 504673.4 447990.4 487701.3 481906.5 484638.3
[5,] 366276.3 376103.3 462953.0 469183.9 414255.2 457252.1 451906.3 454389.7
[6,] 369515.4 379870.1 469230.7 475017.2 420525.0 458361.3 452867.2 455432.0
[,25] [,26] [,27] [,28] [,29] [,30] [,31] [,32]
[1,] 464652.3 299981.1 510752.5 418248.8 505678.9 420970.6 414545.3 511192.3
[2,] 483027.8 330919.8 521202.6 441809.4 517218.3 441358.9 439951.5 521601.9
[3,] 402650.7 291501.4 425730.9 374746.0 423505.7 366566.1 377856.1 426063.2
[4,] 448060.4 308804.8 481527.2 411083.4 478080.6 408051.8 410905.1 481906.5
[5,] 414154.5 272085.1 451503.5 375367.4 447454.6 373286.0 374759.0 451906.3
[6,] 416581.9 277040.4 452471.9 378783.0 448620.7 376131.0 378506.6 452867.2
[,33] [,34] [,35] [,36] [,37] [,38] [,39] [,40]
[1,] 416609.9 418248.8 466325.2 512493.3 418765.5 506313.1 418248.8 416609.9
[2,] 439907.0 441809.4 507210.1 522549.7 442107.7 517823.8 441809.4 439907.0
[3,] 372232.9 374746.0 504835.5 426465.1 374474.5 424056.7 374746.0 372232.9
[4,] 408964.5 411083.4 500652.9 482687.9 411191.1 478670.3 411083.4 408964.5
[5,] 373312.7 375367.4 466999.6 452886.9 375532.7 448061.2 375367.4 373312.7
[6,] 376684.1 378783.0 473277.8 453784.1 378908.8 449221.8 378783.0 376684.1
[,41] [,42] [,43] [,44] [,45] [,46] [,47] [,48]
[1,] 422789.4 414206.8 410709.5 432473.6 520064.2 493167.6 321776.9 134282.591
[2,] 446152.4 434813.9 433243.2 457741.8 530705.7 504484.5 326473.0 116905.672
[3,] 378436.9 360737.2 363872.1 394660.3 435404.2 410601.2 226480.9 5686.274
[4,] 415229.9 401710.3 401694.0 428458.7 491098.6 465269.2 285019.9 73038.156
[5,] 379575.6 366851.1 366234.5 392374.1 460993.8 434727.7 257715.0 69223.198
[6,] 382949.1 369745.5 369479.5 396069.5 461990.6 435862.7 257770.8 63856.791
[,49] [,50] [,51] [,52] [,53] [,54] [,55] [,56]
[1,] 142571.2 446391.6 390185.8 322428.9 583224.2 340912.5 520338.0 416255.0
[2,] 181868.2 465793.9 398746.0 326998.0 588748.4 373017.3 531261.2 439796.5
[3,] 195526.5 388069.2 302508.2 226812.7 486042.8 334930.2 436386.8 372752.7
[4,] 176487.0 431630.6 358570.5 285495.4 546961.8 351878.2 491787.4 409066.0
[5,] 145427.5 397285.5 329207.6 258280.3 520015.9 315196.5 461529.8 373350.1
[6,] 152056.0 399918.7 329926.1 258310.8 520083.6 320236.9 462576.6 376765.5
[,57] [,58] [,59] [,60] [,61] [,62] [,63] [,64]
[1,] 595016.7 417683.1 329530.0 612145.0 424412.6 408594.4 289743.3 416519.3
[2,] 597203.6 443984.0 361276.9 621512.0 444759.8 428336.3 295120.2 439810.7
[3,] 490990.8 384234.7 322617.8 523438.0 369767.5 352474.2 197262.5 372125.4
[4,] 554410.4 415768.8 339804.8 581161.2 411403.2 394614.2 254098.7 408863.9
[5,] 529681.0 379459.5 303107.3 551961.2 376661.6 360032.2 226102.4 373213.4
[6,] 529167.0 383359.1 308115.6 552689.7 379494.1 362779.4 226331.3 376583.9
[,65] [,66] [,67] [,68] [,69] [,70] [,71] [,72]
[1,] 416952.9 410333.6 402240.8 421187.3 422655.2 510575.8 431874.4 415338.0
[2,] 439817.6 450830.6 424863.9 446330.3 443911.8 520959.6 455552.3 441410.1
[3,] 371051.3 447644.8 356013.9 383292.5 371076.7 425390.1 388338.9 381109.3
[4,] 408507.2 443578.7 393441.2 416998.5 411264.4 481253.6 424849.0 412989.1
[5,] 372972.1 409796.0 357933.9 380919.3 376219.5 451266.1 389134.0 376716.8
[6,] 376266.1 416058.1 361208.9 384610.0 379217.6 452222.7 392549.6 380580.1
[,73] [,74] [,75] [,76] [,77] [,78] [,79] [,80]
[1,] 512021.2 330364.5 514631.2 415217.7 437196.7 120824.18 297467.6 535802.3
[2,] 522238.6 362064.6 525092.4 441316.0 454635.9 119074.53 293415.5 541438.3
[3,] 426400.2 323193.1 429587.1 381092.2 372992.6 41594.13 184465.8 439221.1
[4,] 482451.9 340524.7 485413.1 412920.9 419153.1 77925.82 249728.6 499755.5
[5,] 452560.2 303823.1 455393.4 376643.7 385539.7 52026.30 228976.5 472644.2
[6,] 453486.3 308821.6 456361.4 380511.8 387831.3 50845.01 227339.2 472746.4
[,81] [,82] [,83] [,84] [,85] [,86] [,87] [,88]
[1,] 500823.0 383997.5 438936.7 497415.3 418836.9 409309.1 418088.5 399306.2
[2,] 502990.0 396960.4 440393.2 534072.7 441901.9 433241.0 441175.8 421915.0
[3,] 397316.3 308182.8 334549.1 507782.8 373570.8 367440.2 372925.4 353137.4
[4,] 460291.6 359005.2 397597.2 518476.8 410749.1 402893.0 410046.1 390499.2
[5,] 435402.3 327216.3 373051.5 482381.6 375164.3 367063.6 374453.6 354987.7
[6,] 434901.8 328759.7 372438.3 487944.9 378491.0 370559.6 377785.4 358265.2
[,89] [,90] [,91] [,92] [,93] [,94] [,95] [,96]
[1,] 414984.7 399873.8 410174.4 409270.2 411605.2 285790.5 207238.4 506081.8
[2,] 438915.5 422572.2 435056.1 429662.5 436629.5 291830.0 227426.2 517630.0
[3,] 372911.1 353995.5 371711.8 355254.0 373613.4 195156.3 167447.9 423925.8
[4,] 408531.5 391227.4 405553.4 396423.4 407248.3 251102.9 196417.9 478495.7
[5,] 372715.4 355692.9 369508.1 361620.4 371175.3 222637.9 160536.8 447866.1
[6,] 376201.9 358985.3 373170.6 364483.9 374860.6 223005.0 164053.8 449033.5
[,97] [,98] [,99] [,100] [,101] [,102] [,103] [,104]
[1,] 513523.9 342156.5 630527.1 506063.7 515536.1 429635.8 235074.2 520620.7
[2,] 523714.5 374491.3 662528.0 517583.5 525028.5 454531.4 266958.4 530025.2
[3,] 427816.9 337143.1 615179.9 423834.3 428079.4 390549.9 236804.3 432888.5
[4,] 483912.1 353634.9 639648.8 478435.1 484905.7 424922.6 247084.4 489852.5
[5,] 454039.1 316971.8 602897.2 447820.6 455429.8 388913.1 210619.2 460439.5
[6,] 454959.5 322047.3 607561.0 448982.9 456224.8 392547.2 215966.9 461215.9
[,105] [,106] [,107] [,108] [,109] [,110] [,111] [,112]
[1,] 394927.7 414129.8 427831.6 399012.4 409191.3 406292.6 630337.0 416952.9
[2,] 418543.8 438081.6 449113.9 422811.4 429565.2 436473.2 662329.1 439817.6
[3,] 352467.7 372160.9 376178.3 357048.5 355117.3 388791.4 614954.1 371051.3
[4,] 388013.6 407721.1 416458.6 392413.4 396312.8 412440.1 639439.6 408507.2
[5,] 352226.4 371898.0 381419.9 356591.6 361515.6 375700.7 602688.1 372972.1
[6,] 355691.1 375389.6 384415.0 360081.5 364375.9 380290.2 607350.4 376266.1
[,113] [,114] [,115] [,116] [,117] [,118] [,119] [,120]
[1,] 426473.8 119382.28 497802.6 570913.2 415236.0 425087.9 518777.5 420279.0
[2,] 451212.1 144098.59 534546.1 587439.6 438661.8 450709.2 528970.1 440120.2
[3,] 386919.4 120154.44 508633.9 501482.2 371359.9 388828.4 433009.3 364111.0
[4,] 421478.8 121079.19 519094.7 550920.4 407837.8 421799.2 489156.7 406407.4
[5,] 385498.1 84616.82 483024.9 518028.2 372149.2 385629.4 459295.9 371827.8
[6,] 389109.3 90076.99 488602.1 520034.5 375545.7 389398.1 460213.7 374575.4
[,121] [,122] [,123] [,124] [,125] [,126] [,127] [,128]
[1,] 604048.2 340848.2 73500.537 470984.0 412865.9 413454.5 392788.6 413137.1
[2,] 636008.0 372959.5 73190.757 511545.7 434334.9 438259.8 416196.4 433653.7
[3,] 588973.4 334898.9 60132.588 506856.5 362318.3 374594.0 349671.3 359403.6
[4,] 613160.8 351829.3 38508.305 504107.5 401910.1 408666.6 385500.3 400487.1
[5,] 576408.4 315148.2 4054.704 470144.3 366770.9 372644.7 349759.7 365654.6
[6,] 581083.7 320189.9 6363.175 476367.9 369822.1 376288.3 353191.1 368534.4
[,129] [,130] [,131] [,132] [,133] [,134] [,135] [,136]
[1,] 341697.1 413279.4 423761.5 414828.6 424995.4 434088.0 418672.9 406570.3
[2,] 373959.9 433703.6 445789.8 437888.4 443584.3 459372.5 441767.4 425944.4
[3,] 336383.3 359236.7 374754.7 369680.0 364687.6 396282.4 373515.1 349327.7
[4,] 353015.2 400466.0 413751.0 406754.9 408945.0 430095.2 410640.3 391964.1
[5,] 316346.2 365664.8 378472.3 371161.6 374838.6 394010.2 375047.3 357506.0
[6,] 321410.8 368527.7 381606.7 374493.7 377353.7 397706.1 378379.4 360189.9
[,137] [,138] [,139] [,140] [,141] [,142] [,143] [,144]
[1,] 400647.9 429297.9 426780.5 413498.4 236357.9 419164.4 391066.6 428519.3
[2,] 425448.4 454154.7 448038.8 433230.7 268451.7 442526.6 416556.4 449616.1
[3,] 362243.2 390081.6 375078.9 357189.0 238865.6 374930.4 355587.4 376225.6
[4,] 395933.7 424512.6 415370.1 399474.1 248817.4 411624.7 387746.1 416812.1
[5,] 359885.7 388510.9 380336.2 364911.4 212381.4 375962.1 351537.5 381834.6
[6,] 363550.1 392138.7 383328.6 367649.4 217755.2 379341.0 355340.7 384795.5
[,145] [,146] [,147] [,148] [,149] [,150] [,151] [,152]
[1,] 505678.9 382301.6 419133.9 503205.0 424843.1 415085.8 419343.1 418529.5
[2,] 517218.3 406733.7 439088.2 515392.5 445132.7 440455.8 445902.2 441840.6
[3,] 423505.7 343289.4 363368.4 422760.8 369996.1 378243.5 386816.9 374136.7
[4,] 478080.6 377013.8 405465.4 476588.5 411730.3 411372.4 417928.0 410898.7
[5,] 447454.6 341003.9 370842.8 445615.9 377009.6 375234.6 381577.1 375247.8
[6,] 448620.7 344636.9 373612.4 446899.5 379831.1 378975.0 385519.2 378618.7
[,153] [,154] [,155] [,156] [,157]
[1,] 545155.3 419347.1 623743.6 459359.3 413416.5
[2,] 555740.3 442559.7 655380.5 480771.0 433997.3
[3,] 460053.8 374580.8 606903.8 407244.1 359885.8
[4,] 516052.6 411529.0 632069.0 448059.8 400878.2
[5,] 486038.4 375906.1 595324.9 413060.6 366025.2
[6,] 487011.1 379258.2 599930.5 416036.4 368916.1Distance
read_csv("data/US/COP2020/COP2020_tract.txt") %>%
filter(STATEFP == "04") %>%
st_as_sf(coords = c("LONGITUDE", "LATITUDE"), crs = 4326) %>%
mutate(nearest = st_nearest_feature(., AZ_hosp)) %>%
mutate(distance = st_distance(., AZ_hosp[nearest,], by_element = TRUE))Simple feature collection with 1765 features and 6 fields
Geometry type: POINT
Dimension: XY
Bounding box: xmin: -114.7927 ymin: 31.33967 xmax: -109.0779 ymax: 36.98995
Geodetic CRS: WGS 84
# A tibble: 1,765 × 7
STATEFP COUNTYFP TRACTCE POPULATION geometry nearest
* <chr> <chr> <chr> <dbl> <POINT [°]> <int>
1 04 001 942600 1549 (-109.8449 36.76841) 123
2 04 001 942700 4491 (-109.3544 36.77035) 123
3 04 001 944000 5348 (-109.088 35.78314) 48
4 04 001 944100 5495 (-109.2781 36.37803) 123
5 04 001 944201 4021 (-109.6018 36.17521) 123
6 04 001 944202 3608 (-109.5276 36.16716) 123
7 04 001 944301 3789 (-109.7019 36.34859) 123
8 04 001 944302 2685 (-109.6809 36.00098) 123
9 04 001 944901 3369 (-109.4626 35.66766) 78
10 04 001 944902 4201 (-109.7004 35.67758) 78
# ℹ 1,755 more rows
# ℹ 1 more variable: distance [m]Geocoding
- Allows us to retrieve coordinates for addresses
- Interfaces with multiple geocoders
- DO NOT use an non-compliant geocoder for private data
- Geocodio has a paid HIPPA compliant geocoder
- Addresses are sufficient information to reidentify individuals
- We can use the
tidygeocoderpackage to geocode addresses
Geocoding
library(tidygeocoder)
AZ_hosp %>%
mutate(addr =
str_c(ADDRESS, ", ", CITY, ", AZ ", ZIP)) %>%
slice_head(n = 5) %>%
geocode(addr, method = "osm")# A tibble: 5 × 58
OBJECTID OBJECTID_1 RUN_DATE source BUREAU FACID FACILITY_N LICENSE_NU
<int> <int> <date> <chr> <chr> <chr> <chr> <chr>
1 1 2822 2024-05-06 ASPEN MED AZTH00002 BANNER- UNI… <NA>
2 2 2823 2024-05-06 ASPEN MED AZTH00003 BANNER UNIV… <NA>
3 3 2824 2024-05-06 ASPEN MED AZTH00004 MAYO CLINIC… <NA>
4 4 3774 2024-05-06 ASPEN MED BH7347 VIA LINDA B… SH11520
5 5 4893 2024-05-06 ASPEN MED MED0198 CANYON VIST… H7130
# ℹ 50 more variables: LICENSE_EF <date>, LICENSE_EX <date>, MEDICARE_I <chr>,
# MEDICARE_1 <chr>, MEDICARE_2 <chr>, TELEPHONE <chr>, FACILITY_T <int>,
# TYPE <chr>, SUBTYPE <chr>, CATEGORY <chr>, ICON_CATEG <chr>,
# MEDICARE_T <chr>, LICENSE_TY <chr>, LICENSE_SU <chr>, CAPACITY <int>,
# ADDRESS <chr>, CITY <chr>, ZIP <int>, COUNTY <chr>, OPERATION_ <chr>,
# X_Is_Publi <chr>, HOSPITAL_G <chr>, KeyID <chr>, ADHSCODE <chr>,
# N_AddressT <chr>, N_LocatorT <chr>, N_ADDRESS <chr>, N_ADDR2 <chr>, …Best Practices
Map making is an art
- Good map making takes into consideration the
- audience
- medium
- message

Section 508 Compliance
- Make your maps accessible by
- Using a monochromatic color scale (unless appropriate)
- Providing descriptive text or alt text
- If possible, including map, description, and data together
- Using a monochromatic color scale (unless appropriate)
Be Careful With Location Data
- Locations are often sensitive
- You can mask locations using
- Symbol maps
- Dot density maps
- Converting to rates and using a choropleth map
Provide Metadata
It is a good practice to add the following information to your map:
- Author
- Date
- Data source and year
Stop and make some maps
- Use a table join to join the Area Deprivation Index to block groups within Arizona. Create a map of Area Deprivation Index at the block group level for the state of Arizona
Stop and make some maps
- Create a graduated symbol map of the number of WIC Vendors within Arizona counties using quantile breaks. Using
cowplot, place a quantile breaks choropleth map of diabetes adjusted prevalence (PLACES) also within Arizona counties.
Stop and make some maps
- Create a continuous choropleth map of distance to the nearest pharmacy at the block group level for Maricopa County
Stop and make some maps
- Estimate the number of individuals within 15 miles of a hospital.
- Create a buffer around the hospitals
- Convert the centers of population block group table to an
sfobject - Spatially join the centers of population to the hospital buffers
- Filter out centers of population not within a buffer, and sum the population

